A mathematical model of H5N1 influenza transmission in US dairy cattle
- PMID: 40341525
- PMCID: PMC12062519
- DOI: 10.1038/s41467-025-59554-z
A mathematical model of H5N1 influenza transmission in US dairy cattle
Abstract
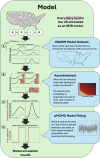
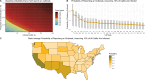
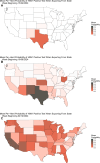
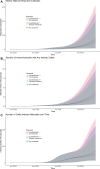
2024 saw a novel outbreak of H5N1 avian influenza in US dairy cattle. Limited surveillance data has made determining the true scale of the epidemic difficult. We present a stochastic metapopulation transmission model that simulates H5N1 influenza transmission through individual dairy cows in 35,974 herds in the continental US. Transmission is enabled through the movement of cattle between herds, as indicated from Interstate Certificates of Veterinary Inspection data. We estimate the rates of under-reporting by state and present the anticipated rates of positivity for cattle tested at the point of exportation over time. We investigate the impact of intervention methods on the underlying epidemiological dynamics, demonstrating that current interventions have had insufficient impact, preventing only a mean 175.2 reported outbreaks. Our model predicts that the majority of the disease burden is, as of January 2025, concentrated within West Coast states. We quantify the uncertainty in the scale of the epidemic, highlighting the most pressing data streams to capture, and which states are expected to see outbreaks emerge next, with Arizona and Wisconsin at greatest risk. Our model suggests that dairy outbreaks will continue to occur in 2025, and that more urgent, farm-focused, biosecurity interventions and targeted surveillance schemes are needed.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: We declare that none of the authors have competing financial or non-financial interests as defined by Nature Portfolio. Inclusion and Ethics: All roles and responsibilities were agreed amongst collaborators ahead of the research. Local ethics review was not required due to this being a computational study.
Figures

References
-
- Klein, B., Kraemer, M. U. G. & Scarpino, S. V. Timeline for H5N1 in the USA during the 2024 Outbreak. https://github.com/Emergent-Epidemics/H5N1_US2024_timeline (2024).
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

