Targeted disruption of PRC1.1 complex enhances bone remodeling
- PMID: 40341537
- PMCID: PMC12062457
- DOI: 10.1038/s41467-025-59638-w
Targeted disruption of PRC1.1 complex enhances bone remodeling
Abstract
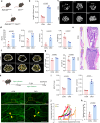
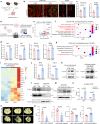
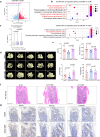
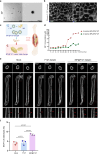
Polycomb repressive complexes (PRCs) are pivotal epigenetic regulators that preserve cell identity by restricting transcription responses to sub-threshold extracellular signals. Their roles in osteoblast function and bone formation remain unclear. Here in aging osteoblasts, we found marked activation of PRC1.1 complex, with KDM2B acting as a chromatin-binding factor and BCOR and PCGF1 enabling histone H2A monoubiquitylation (H2AK119ub1). Osteoblast-specific Kdm2b inactivation significantly enhances bone remodeling under steady-state conditions and in scenarios of bone loss. This enhancement is attributed to H2AK119ub1 downregulation and subsequent Wnt signaling derepression. Furthermore, we developed a small molecule termed iBP, that specifically inhibits the interaction between BCOR and PCGF1, thereby suppressing PRC1.1 activity. Notably, iBP administration promotes bone formation in mouse models of bone loss. Therefore, our findings identify PRC1.1 as a critical epigenetic brake on bone formation and demonstrate that therapeutic targeting of this complex enhances Wnt pathway activation, offering a promising strategy against skeletal deterioration.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Ethics statement: We support inclusive, diverse, and equitable conduct of research.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

