Comparative analysis of virulence-associated genes in ESBL-producing Escherichia coli isolates from bloodstream and urinary tract infections
- PMID: 40342604
- PMCID: PMC12058657
- DOI: 10.3389/fmicb.2025.1571121
Comparative analysis of virulence-associated genes in ESBL-producing Escherichia coli isolates from bloodstream and urinary tract infections
Abstract
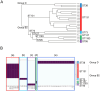
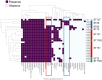
The prevalence of extended-spectrum β-lactamase (ESBL)-producing Escherichia coli (E. coli) is a global health concern due to the multidrug antimicrobial resistance in extraintestinal pathogenic E. coli (ExPEC). ExPEC causes severe infections such as bloodstream infections, meningitis, and sepsis. Uropathogenic E. coli (UPEC), a subset of ExPEC, is responsible for urinary tract infections (UTIs), ranging from asymptomatic bacteriuria and cystitis to more severe conditions, such as pyelonephritis, bacteremia, and sepsis (urosepsis). Although ESBL-producing E. coli may have a significant impact on patient outcomes, comparisons of genotype and virulence factors between ESBL-producing and non-ESBL-producing E. coli have not fully elucidated the factors influencing its pathogenicity. Therefore, in the present study, we analyzed the genotypes and virulence-associated genes of ESBL-producing strains isolated from the blood of patients with UTIs to determine the characteristics of ESBL-producing UPEC strains associated with severe infections. Most of the clinical isolates belonged to phylogroup B2, with the exception of three strains from phylogroup D. The MLST was ST131, followed by ST73, ST95, and ST38, which are commonly found in UPEC strains. Intriguingly, ST131 strains were associated with fewer sepsis cases compared to non-ST131 strains (8 of 38 cases by ST131 and 5 of 8 cases by non-ST131 [OR, 0.16; 95% CI, 0.038-0.873; p = 0.031]). In silico analysis of 23 clinical isolates revealed that the genes detected in all strains may play a significant role in the pathogenesis of invasive UTIs. Clustering and gene locus analysis highlighted the genotype-MLST dependence of UPEC-specific virulence-associated genes. ST38-specific strains were atypical, characterized by the absence of several UPEC-specific genes, including pap loci, pathogenicity island marker (malX), and ompT, as well as the presence of genes encoding Ycb fimbriae and a Type 3 secretion system, which are typically found in enteropathogenic E. coli (EPEC). These results suggest that the virulence of clinical isolates causing invasive infections can vary, and that the pathogenicity of UPEC should be considered when analyzing the correlation between MLST and the repertoire of virulence-associated genes.
Keywords: ESBL-producing Escherichia coli; ST131; bloodstream infections; extraintestinal pathogenic Escherichia coli; uropathogenic Escherichia coli; urosepsis; virulence-associated genes.
Copyright © 2025 Tanaka, Hanawa, Suda, Tanji, Minh, Kondo, Azam, Kiga, Yonetani, Yashiro, Ohmori and Matsuda.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Comparative Genomic Analysis of Globally Dominant ST131 Clone with Other Epidemiologically Successful Extraintestinal Pathogenic Escherichia coli (ExPEC) Lineages.mBio. 2017 Oct 24;8(5):e01596-17. doi: 10.1128/mBio.01596-17. mBio. 2017. PMID: 29066550 Free PMC article.
-
Comparative Genomics of Escherichia coli Isolated from Skin and Soft Tissue and Other Extraintestinal Infections.mBio. 2017 Aug 15;8(4):e01070-17. doi: 10.1128/mBio.01070-17. mBio. 2017. PMID: 28811343 Free PMC article.
-
Genotypic characteristics of Uropathogenic Escherichia coli isolated from complicated urinary tract infection (cUTI) and asymptomatic bacteriuria-a relational analysis.PeerJ. 2023 Jun 20;11:e15305. doi: 10.7717/peerj.15305. eCollection 2023. PeerJ. 2023. PMID: 37361034 Free PMC article.
-
First detection of VEB-1 extended-spectrum β-lactamase-producing Escherichia coli clinical isolate in Japan.Microbiol Spectr. 2024 Nov 5;12(11):e0052324. doi: 10.1128/spectrum.00523-24. Epub 2024 Sep 17. Microbiol Spectr. 2024. PMID: 39287461 Free PMC article.
-
Multidrug resistance dissemination by extended-spectrum β-lactamase-producing Escherichia coli causing community-acquired urinary tract infection in the Central-Western Region, Brazil.J Glob Antimicrob Resist. 2016 Sep;6:1-4. doi: 10.1016/j.jgar.2016.02.003. Epub 2016 Mar 26. J Glob Antimicrob Resist. 2016. PMID: 27530830
References
-
- Abe C. M., Salvador F. A., Falsetti I. N., Vieira M. A. M., Blanco J., Blanco J. E., et al. . (2008). Uropathogenic Escherichia coli (UPEC) strains may carry virulence properties of diarrhoeagenic E. coli. FEMS Immunol. Med. Microbiol. 52, 397–406. doi: 10.1111/j.1574-695X.2008.00388.x, PMID: - DOI - PubMed
-
- Adams-Sapper S., Diep B. A., Perdreau-Remington F., Riley L. W. (2013). Clonal composition and community clustering of drug-susceptible and -resistant Escherichia coli isolates from bloodstream infections. Antimicrob. Agents Chemother. 57, 490–497. doi: 10.1128/AAC.01025-12, PMID: - DOI - PMC - PubMed
-
- Biggel M., Xavier B. B., Johnson J. R., Nielsen K. L., Frimodt-Møller N., Matheeussen V., et al. . (2020). Horizontally acquired papGII-containing pathogenicity islands underlie the emergence of invasive uropathogenic Escherichia coli lineages. Nat. Commun. 11:5968. doi: 10.1038/s41467-020-19714-9, PMID: - DOI - PMC - PubMed
-
- CDC (2019). Antibiotic resistance threats in the United States, 2019. Atlanta, Georgia: U.S. Department of Health and Human Services.
LinkOut - more resources
Full Text Sources
Research Materials

