PITX1 as a grading, prognostic and tumor-infiltrating immune cells marker for chondrosarcoma: a public database-based immunoassay and tissue sample analysis
- PMID: 40342824
- PMCID: PMC12060168
- DOI: 10.3389/fonc.2025.1477649
PITX1 as a grading, prognostic and tumor-infiltrating immune cells marker for chondrosarcoma: a public database-based immunoassay and tissue sample analysis
Abstract
Background: Chondrosarcoma (CHS) is a rare bone cancer originating from chondrocytes, with high-grade cases associated with high mortality rates. However, the prognostic factors and therapeutic targets for CHS have not been studied.
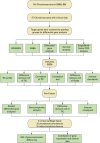
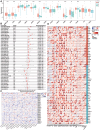
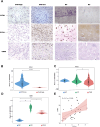
Methods: Graded gene differential analysis was conducted on 97 CHS tissues to identify genes associated with CHS grading. Additionally, we performed GO and KEGG enrichment analyses of the differentially-expressed genes (DEGs), as well as GSEA analysis, differential expression analysis, survival analysis, and univariable and multifactorial COX analysis of paired-like homology structural domain transcription factor 1 (PITX1). Furthermore, our findings investigated the relationship between tumor-infiltrating immune cells (TICs) in CHS tumors using CIBERSORT to calculate proportions and differences. Our findings also explored the associations among gene expression patterns, survival prognosis, TICs, and immune checkpoints across various cancer types. Finally, immunohistochemical staining was carried out on self-collected clinical samples to assess PITX1 expression levels and correlate them with clinical information.
Results: Gene differential expression analysis revealed a strong correlation between PITX1 expression and tumor grade. GO, KEGG enrichment, and GSEA analysis demonstrated the association of PITX1 with cell proliferation-related processes, such as cell cycle regulation and mitosis, and differentiation-related processes, such as RNA processing. PITX1 expression was associated with tumor stage and survival outcomes. Immunoassay indicated a positive correlation between PITX1 levels and TICs, immune checkpoints, and graded TICs. Pan-cancer analysis confirmed the differential expression of the PITX1 gene across multiple cancers, impacting survival prognosis, TIC patterns, and immune checkpoint regulation. Lastly, our 75 collection of clinical patient tissue samples exhibited varying levels of PITX1 expression across different cancer grades while also demonstrating a significant association with tumor differentiation and metastasis.
Conclusion: PITX1 is a novel biomarker for distinguishing between high-grade and low-grade CHS, serving as a prognostic indicator for patients with this condition and presenting a promising target for immunotherapy. These findings offer innovative insights into the treatment of CHS.
Keywords: PITX1; chondrosarcoma (CHS); immune targets; tumor grade; tumor-infiltrating immune cell (TIC).
Copyright © 2025 Huang, Liu, Zhang, Lu, Hu, Zhang, Xie and Chen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources

