RNA splicing modulator for Huntington's disease treatment induces peripheral neuropathy
- PMID: 40343270
- PMCID: PMC12059699
- DOI: 10.1016/j.isci.2025.112380
RNA splicing modulator for Huntington's disease treatment induces peripheral neuropathy
Abstract
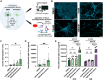
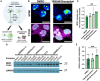
RNA splicing modulators, a new class of small molecules with the potential to modify the protein expression levels, have quickly been translated into clinical trials. These compounds hold promise for treating neurodegenerative disorders, including branaplam for lowering huntingtin levels in Huntington's disease. However, the VIBRANT-HD trial was terminated due to the emergence of peripheral neuropathy. Here, we describe the complex mechanism whereby branaplam activates p53, induces nucleolar stress in human induced pluripotent stem cell (iPSC)-derived motor neurons (iPSC-MN), and thereby enhanced expression of the neurotoxic p53-target gene BBC3. On the cellular level, branaplam disrupts neurite integrity, reflected by elevated neurofilament light chain levels. These findings illustrate the complex pharmacology of RNA splicing modulators with a small therapeutic window between lowering huntingtin levels and the clinically relevant off-target effect of neuropathy. Comprehensive toxicological screening in human stem cell models can complement pre-clinical testing before advancing RNA-targeting drugs to clinical trials.
Keywords: Biological sciences; Cell biology; Cellular neuroscience; Natural sciences; Neuroscience; Pharmacology.
© 2025 The Authors.
Conflict of interest statement
Full financial disclosures of all authors for the previous 12 months. F.K.: Employment (University Hospital Erlangen); T.B.: Employment (University Hospital Erlangen); S.N.: Employment (University Hospital Erlangen); L.K.: Employment (University Hospital Erlangen); M.R.: Employment (University Hospital Erlangen, Klinikum Forchheim), grants (BMBF, Förderverein für HSP-Forschung), honoraria (Orphalan, Desitin, Ever Pharma, Zambon, Bial, DGN), prize (Euro-HSP).; J.W.: Employment (University Hospital Erlangen), honoraria: Zambon, Bial, UCB.; B.W.: Employment (University Hospital Erlangen).
Figures


References
-
- Keller C.G., Shin Y., Monteys A.M., Renaud N., Beibel M., Teider N., Peters T., Faller T., St-Cyr S., Knehr J., et al. An orally available, brain penetrant, small molecule lowers huntingtin levels by enhancing pseudoexon inclusion. Nat. Commun. 2022;13:1150. doi: 10.1038/s41467-022-28653-6. - DOI - PMC - PubMed
-
- Krach F., Stemick J., Boerstler T., Weiss A., Lingos I., Reischl S., Meixner H., Ploetz S., Farrell M., Hehr U., et al. An alternative splicing modulator decreases mutant HTT and improves the molecular fingerprint in Huntington’s disease patient neurons. Nat. Commun. 2022;13:6797. doi: 10.1038/s41467-022-34419-x. - DOI - PMC - PubMed
-
- Branaplam : VIBRANT-HD Study Update Novartis. https://www.novartis.com/news/branaplam-vibrant-hd-study-update
-
- Krach F., Wheeler E.C., Regensburger M., Boerstler T., Wend H., Vu A.Q., Wang R., Reischl S., Boldt K., Batra R., et al. Aberrant NOVA1 function disrupts alternative splicing in early stages of amyotrophic lateral sclerosis. Acta Neuropathol. 2022;144:413–435. doi: 10.1007/s00401-022-02450-3. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

