Plasmodium falciparum isolates: ex vivo drug response
- PMID: 40343740
- PMCID: PMC12209805
- DOI: 10.1093/jac/dkaf129
Plasmodium falciparum isolates: ex vivo drug response
Abstract
Objectives: While artemisinin-based combination therapies (ACTs) are effective in sub-Saharan Africa, clinical isolates that are refractory to artemisinin derivatives are emerging in East Africa and ACT partner drugs are becoming less effective in West Africa. We investigated the ex vivo responses of Plasmodium falciparum clinical isolates to frontline antimalarials and the contribution of validated molecular markers of antimalarial drug resistance.
Methods: Ex vivo susceptibility was measured for 66 clinical isolates collected from uncomplicated malaria patients. IC50 was measured for dihydroartemisinin, artesunate, lumefantrine, amodiaquine and chloroquine using a SYBR Green I growth inhibition assay. We also assessed known drug resistance-mediating polymorphisms in pfcrt, pfmdr1 and pfkelch13 using Oxford Nanopore amplicon sequencing.
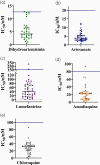
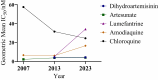
Results: P. falciparum clinical isolates were susceptible to dihydroartemisinin and artesunate. Clinical isolates showed a wide distribution of susceptibility to lumefantrine and amodiaquine, with some parasites having IC50 values above reference cut-offs for resistance to lumefantrine (150 nM) and amodiaquine (60 nM), suggesting decreased drug susceptibility. Ninety-seven percent of the isolates carried WT pfcrt K76 and pfmdr1 N86 alleles, reported to mediate reduced response to lumefantrine and artemether/lumefantrine. pfmdr1 N86 and 184F haplotype was carried by 62.1% of parasites. None of the clinical isolates carried validated pfkelch13 mutations known to mediate artemisinin partial resistance.
Conclusions: Clinical isolates from coastal Ghana remain susceptible to artemisinin derivatives in commonly used ACTs in Ghana. However, we observed lower susceptibility to the ACT partner drugs lumefantrine and amodiaquine, suggesting the emergence of drug-tolerance phenotypes. Consistent surveillance of drug phenotype-genotype is needed to support ACT efficacy in Ghana.
© The Author(s) 2025. Published by Oxford University Press on behalf of British Society for Antimicrobial Chemotherapy.
Figures


References
-
- WHO . World Malaria Report 2022. 2023. https://www.who.int/teams/global-malaria-programme/reports/world-malaria....
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

