Kinome screening identifies integrated stress response kinase EIF2AK1/HRI as a negative regulator of PINK1 mitophagy signaling
- PMID: 40344059
- PMCID: PMC12063660
- DOI: 10.1126/sciadv.adn2528
Kinome screening identifies integrated stress response kinase EIF2AK1/HRI as a negative regulator of PINK1 mitophagy signaling
Abstract
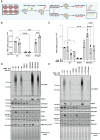
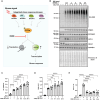
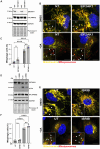
Loss-of-function mutations in the PINK1 kinase lead to early-onset Parkinson's disease (PD). PINK1 is activated by mitochondrial damage to phosphorylate ubiquitin and Parkin, triggering mitophagy. PINK1 also indirectly phosphorylates Rab GTPases, such as Rab8A. Using an siRNA library targeting human Ser/Thr kinases in HeLa cells, we identified EIF2AK1 [heme-regulated inhibitor (HRI) kinase], a branch of the integrated stress response (ISR), as a negative regulator of PINK1. EIF2AK1 knockdown enhances mitochondrial depolarization-induced PINK1 stabilization and phosphorylation of ubiquitin and Rab8A. These results were confirmed in SK-OV-3, U2OS, and ARPE-19 cells. Knockdown of DELE1, an activator of EIF2AK1, produced similar effects. Notably, the ISR inhibitor ISRIB also enhanced PINK1 activation. In human cells with mito-QC mitophagy reporters, EIF2AK1 knockdown or ISRIB treatment increased PINK1-dependent mitophagy without affecting deferiprone-induced mitophagy. These findings suggest that the DELE1-EIF2AK1 ISR pathway is a negative regulator of PINK1-dependent mitophagy. Further evaluation in PD-relevant models is needed to assess the therapeutic potential of targeting this pathway.
Figures




References
-
- Kitada T., Asakawa S., Hattori N., Matsumine H., Yamamura Y., Minoshima S., Yokochi M., Mizuno Y., Shimizu N., Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 392, 605–608 (1998). - PubMed
-
- Valente E. M., Abou-Sleiman P. M., Caputo V., Muqit M. M., Harvey K., Gispert S., Ali Z., Del Turco D., Bentivoglio A. R., Healy D. G., Albanese A., Nussbaum R., Gonzalez-Maldonado R., Deller T., Salvi S., Cortelli P., Gilks W. P., Latchman D. S., Harvey R. J., Dallapiccola B., Auburger G., Wood N. W., Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science 304, 1158–1160 (2004). - PubMed
-
- Kakade P., Ojha H., Raimi O. G., Shaw A., Waddell A. D., Ault J. R., Burel S., Brockmann K., Kumar A., Ahangar M. S., Krysztofinska E. M., Macartney T., Bayliss R., Fitzgerald J. C., Muqit M. M. K., Mapping of a N-terminal α-helix domain required for human PINK1 stabilization, Serine228 autophosphorylation and activation in cells. Open Biol. 12, 210264 (2022). - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

