Evolution of primate T-cell leukemia virus type 1 accessory genes and functional divergence of its antisense proteins
- PMID: 40344170
- PMCID: PMC12088518
- DOI: 10.1371/journal.ppat.1013158
Evolution of primate T-cell leukemia virus type 1 accessory genes and functional divergence of its antisense proteins
Erratum in
-
Correction: Evolution of primate T-cell leukemia virus type 1 accessory genes and functional divergence of its antisense proteins.PLoS Pathog. 2025 Nov 7;21(11):e1013668. doi: 10.1371/journal.ppat.1013668. eCollection 2025 Nov. PLoS Pathog. 2025. PMID: 41201998 Free PMC article.
Abstract
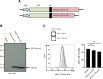
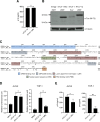
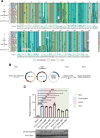
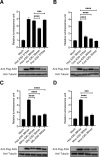
Human T-cell leukemia virus type 1 (HTLV-1) is derived from simian T-cell leukemia virus type 1 (STLV-1), and together they form a broader category known as primate T-cell leukemia virus type 1 (PTLV-1). PTLV-1 encodes multiple proteins from overlapping open reading frames (ORFs) in the pX region. This study aims to characterize the conservation of these proteins in different PTLV-1 subtypes and their role in pathogenesis. For the first time, we report the full-length proviral sequence of an STLV-1 strain isolated from chimpanzee and African green monkey. Phylogenetic analysis reveals high conservation of the accessory proteins p12, p30, and p13 in the HTLV-1a subtype. Conversely, some African PTLV-1 subtypes exhibit loss of ORFs for p12 or p13. For Asian subtypes, simian strains often lack p12, p13, or p30 proteins, whereas human strains retain the ORFs of p30 and p13 but not p12. To assess the infectivity of a simian strain of PTLV-1 lacking ORFs for p12, p13, and p30, we constructed a molecular clone from a naturally infected Japanese macaque (Mfu: Macaca fuscata) and compared it with HTLV-1a. Using a reporter assay and ELISA, we found similar infectivity to Jurkat T cells; however, STLV-1 Mfu exhibited impaired infectivity in the monocytic cell line THP-1. Additionally, despite the conservation of the HTLV-1/STLV-1 bZIP factor (HBZ/SBZ) ORFs, HBZ/SBZ proteins derived from HTLV-1a and African PTLV-1 subtypes induce significantly higher activation of the TGF-β/Smad signaling pathway than those from Asian subtypes. Collectively, our findings suggest that the acquisition of the accessory proteins by PTLV-1 subtypes potentially confers an advantageous adaptation of PTLV-1 during infection in apes, including humans. Moreover, among PTLV-1 strains, HBZ/SBZ had varying degrees of activity on the TGF-β/Smad pathway; this fact underscores the complex interplay between viral proteins and host signaling pathways, possibly influencing the viral pathogenicity in different species.
Copyright: © 2025 Hussein et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist
Figures



References
-
- Matsuoka M, Jeang KT. Human T-cell leukaemia virus type 1 (HTLV-1) infectivity and cellular transformation. Nat Rev Cancer. 2007;7:270–80. - PubMed
-
- Akari H, Ono F, Sakakibara I, Takahashi H, Murayama Y, Hiyaoka A, et al. Simian T cell leukemia virus type I-induced malignant adult T cell leukemia-like disease in a naturally infected African green monkey: implication of CD8+ T cell leukemia. AIDS Res Hum Retroviruses. 1998;14(4):367–71. doi: 10.1089/aid.1998.14.367 - DOI - PubMed
-
- Tsujimoto H, Noda Y, Ishikawa K, Nakamura H, Fukasawa M, Sakakibara I, et al. Development of adult T-cell leukemia-like disease in African green monkey associated with clonal integration of simian T-cell leukemia virus type I. Cancer Res. 1987;47(1):269–74. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

