Msp1 and Pex19-Pex3 cooperate to achieve correct localization of Pex15 to peroxisomes
- PMID: 40344504
- PMCID: PMC12366275
- DOI: 10.1111/febs.70132
Msp1 and Pex19-Pex3 cooperate to achieve correct localization of Pex15 to peroxisomes
Abstract
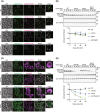
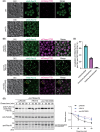
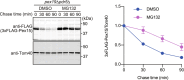
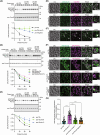
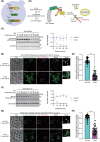
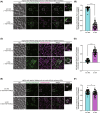
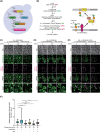
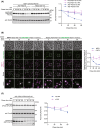
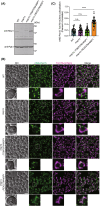
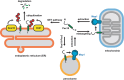
Yeast Msp1 is a dual-localized AAA-ATPase on the mitochondrial outer membrane (OM) and peroxisomal membrane. We previously showed that Msp1 transfers mistargeted tail-anchored (TA) proteins from mitochondria to the endoplasmic reticulum (ER) for degradation or delivery to their original destinations. However, the mechanism by which Msp1 in mitochondria and peroxisomes handles authentic peroxisomal TA proteins remains unclear. We show that newly synthesized Pex15 is targeted to peroxisomes primarily via the Pex19- and Pex3-dependent pathway. Mistargeted Pex15 on the mitochondrial OM is extracted by mitochondrial Msp1 and transferred to the ER via the guided-entry of TA proteins pathway for degradation or to peroxisomes via the Pex19-Pex3 pathway. Intriguingly, endogenous Pex15 localized in peroxisomes is also extracted from the membranes by peroxisomal Msp1 but returns to peroxisomes via the Pex19-Pex3 pathway. These results suggest that correct Pex15 localization to peroxisomes relies on not only the initial targeting by Pex19-Pex3 but also the constant re-routing by Msp1 and Pex19-Pex3.
Keywords: Msp1; Pex15; Pex19‐Pex3; mitochondria; peroxisome.
© 2025 The Author(s). The FEBS Journal published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Wattenberg B & Lithgow T (2001) Targeting of C‐terminal (tail)‐anchored proteins: understanding how cytoplasmic activities are anchored to intracellular membranes. Traffic 2, 66–71. - PubMed
-
- Wang L & Walter P (2020) Msp1/ATAD1 in protein quality control and regulation of synaptic activities. Annu Rev Cell Dev Biol 36, 141–164. - PubMed
-
- Borgese N & Fasana E (2011) Targeting pathways of C‐tail‐anchored proteins. Biochim Biophys Acta 2808, 937–946. - PubMed
MeSH terms
Substances
Grants and funding
- Foundation of Kinoshita Memorial Enterprise
- Japan Foundation for Applied Enzymology
- 15H05705/Japan Society for the Promotion of Science
- 20H00458/Japan Society for the Promotion of Science
- 20H04912/Japan Society for the Promotion of Science
- 20H05689/Japan Society for the Promotion of Science
- 20H05929/Japan Society for the Promotion of Science
- 23H02437/Japan Society for the Promotion of Science
- 23K17995/Japan Society for the Promotion of Science
- 24K18117/Japan Society for the Promotion of Science
- 24KJ0210/Japan Society for the Promotion of Science
- Takeda Science Foundation
- 21gm1410002/Japan Agency for Medical Research and Development
LinkOut - more resources
Full Text Sources

