Disentangling the Switching Behavior in Functional Connectivity Dynamics in Autism Spectrum Disorder: Insights from Developmental Cohort Analysis and Molecular-Cellular Associations
- PMID: 40344520
- PMCID: PMC12120798
- DOI: 10.1002/advs.202403801
Disentangling the Switching Behavior in Functional Connectivity Dynamics in Autism Spectrum Disorder: Insights from Developmental Cohort Analysis and Molecular-Cellular Associations
Abstract
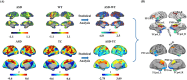
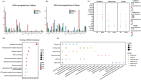
Characterizing the transition or switching behavior between multistable brain states in functional connectivity dynamics (FCD) holds promise for uncovering the underlying neuropathology of Autism Spectrum Disorder (ASD). However, whether and how switching behaviors in FCD change in patients with developmental ASD, as well as their cellular and molecular basis, remains unexplored. This study develops a region-wise FCD switching index (RFSI) to investigate the drivers of FCD. This work finds that brain regions within the salience, default mode, and frontoparietal networks serve as abnormal drivers of FCD in ASD across different developmental stages. Additionally, changes in RFSI at different developmental stages of ASD correlated with transcriptomic profiles and neurotransmitter density maps. Importantly, the abnormal RFSI identifies in humans has also been observed in genetically edited ASD monkeys. Finally, single-nucleus RNA sequencing data from patients with developmental ASD are analyzed and aberrant switching behaviors in FCD may be mediated by somatostatin-expressing interneurons and altered differentiation patterns in astrocyte State2. In conclusion, this study provides the first evidence of abnormal drivers of FCD across different stages of ASD and their associated cellular and molecular mechanisms. These findings deepen the understanding of ASD neuropathology and offer valuable insights into treatment strategies.
Keywords: autism spectrum disorder; brain dynamics; development; molecular mechanisms; resting‐state fMRI.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Dynamic functional connectivity analysis reveals decreased variability of the default-mode network in developing autistic brain.Autism Res. 2018 Nov;11(11):1479-1493. doi: 10.1002/aur.2020. Epub 2018 Oct 1. Autism Res. 2018. PMID: 30270547
-
Developmental changes in large-scale network connectivity in autism.Neuroimage Clin. 2015 Mar 6;7:732-41. doi: 10.1016/j.nicl.2015.02.024. eCollection 2015. Neuroimage Clin. 2015. PMID: 25844325 Free PMC article.
-
Developmental functional brain network abnormalities in autism spectrum disorder comorbid with attention deficit hyperactivity disorder.Eur J Pediatr. 2025 Jan 31;184(2):166. doi: 10.1007/s00431-025-05989-x. Eur J Pediatr. 2025. PMID: 39888443
-
Atypical longitudinal development of functional connectivity in adolescents with autism spectrum disorder.Autism Res. 2019 Jan;12(1):53-65. doi: 10.1002/aur.1971. Epub 2018 Oct 30. Autism Res. 2019. PMID: 30375176 Free PMC article.
-
Alternative splicing in autism spectrum disorder: Recent insights from mechanisms to therapy.Asian J Psychiatr. 2025 Jun;108:104501. doi: 10.1016/j.ajp.2025.104501. Epub 2025 Apr 14. Asian J Psychiatr. 2025. PMID: 40273800 Review.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
