DDX3 Regulates the Cap-Independent Translation of the Japanese Encephalitis Virus via Its Interactions with PABP1 and the Untranslated Regions of the Viral Genome
- PMID: 40344524
- PMCID: PMC12279179
- DOI: 10.1002/advs.202502493
DDX3 Regulates the Cap-Independent Translation of the Japanese Encephalitis Virus via Its Interactions with PABP1 and the Untranslated Regions of the Viral Genome
Abstract
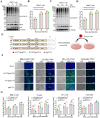
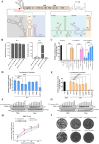
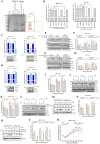
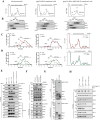
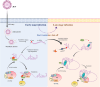
The translation of global cellular proteins is almost completely repressed in cells with flavivirus infection, while viral translation remains efficient. The mechanisms of flaviviruses evade host translational shutoff are largely unknown. Here, it is found that Japanese encephalitis virus (JEV) can adopt cap-independent (CI) translation to escape the host translational shutoff. Furthermore, the elements DB2 and sHP-SL within 3'UTR are involved in the regulation of CI translation, which is conserved in the genus Orthoflavivirus. By RNA affinity purification and mass spectrometry analysis, cellular DEAD-box protein 3 (DDX3) and poly(A)-binding protein 1 (PABP1) are identified as key factors in regulating CI translation of JEV via their interactions with DB2 and sHP-SL RNA structures. Mechanistically, it is revealed that DDX3 binds to both 5'UTR and 3'UTR of the JEV genome to establish a closed-loop architecture and recruit eIF4G/eIF4A to form the DDX3/PABP1/eIF4G/eIF4A tetrameric complex via its interaction with PABP1, thereby recruiting the ribosomal 43S preinitiation complex (PIC) to the 5'-end of the JEV genome to start translation. These findings demonstrate a noncanonical translation strategy employed by JEV and further reveal the regulatory roles of DDX3 and PABP1 in this mechanism. These results expand the knowledge of the translation initiation regulation in flaviviruses under the state of host translational shutoff, which provides a conserved antiviral target against orthoflavivirus.
Keywords: DDX3; Japanese encephalitis virus; cap‐independent translation.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Cellular NONO protein binds to the flavivirus replication complex and promotes positive-strand RNA synthesis.J Virol. 2024 Dec 17;98(12):e0029724. doi: 10.1128/jvi.00297-24. Epub 2024 Nov 5. J Virol. 2024. PMID: 39499073 Free PMC article.
-
Cellular DDX3 regulates Japanese encephalitis virus replication by interacting with viral un-translated regions.Virology. 2014 Jan 20;449:70-81. doi: 10.1016/j.virol.2013.11.008. Epub 2013 Nov 26. Virology. 2014. PMID: 24418539 Free PMC article.
-
Interaction of West Nile virus NS5 with orthoflavivirus SLA RNAs and their effects on viral replication and inhibition.J Virol. 2025 Aug 18:e0202324. doi: 10.1128/jvi.02023-24. Online ahead of print. J Virol. 2025. PMID: 40824093
-
Meta-analyses of the proportion of Japanese encephalitis virus infection in vectors and vertebrate hosts.Parasit Vectors. 2017 Sep 7;10(1):418. doi: 10.1186/s13071-017-2354-7. Parasit Vectors. 2017. PMID: 28882172 Free PMC article.
-
Assessment of data on vector and host competence for Japanese encephalitis virus: A systematic review of the literature.Prev Vet Med. 2018 Jun 1;154:71-89. doi: 10.1016/j.prevetmed.2018.03.018. Epub 2018 Mar 22. Prev Vet Med. 2018. PMID: 29685447
References
-
- Yamada M., Nakamura K., Yoshii M., Kaku Y., Narita M., J. Comp. Pathol. 2009, 141, 156. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
