Targeting Stat3 with conditional knockout or PROTAC technology alleviates renal injury by Limiting pyroptosis
- PMID: 40344718
- PMCID: PMC12136849
- DOI: 10.1016/j.ebiom.2025.105739
Targeting Stat3 with conditional knockout or PROTAC technology alleviates renal injury by Limiting pyroptosis
Abstract
Background: Acute kidney injury (AKI) is a critical clinical syndrome with high morbidity, mortality, and no effective treatment in clinical practice. The role of the Signal Transducer and Activator of Transcription 3 (Stat3) in AKI remains controversial, and its complex regulatory mechanisms must be further explored.
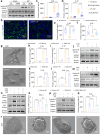
Methods: We generated renal tubular epithelial cells Stat3 conditional knockout (cKO) mice and used them in cecal ligation and puncture (CLP) and ischaemia-reperfusion (I/R) induced AKI models. Additionally, proteolysis-targeting chimaera (PROTAC) compound E034 was designed and synthesised. We also utilised human kidney tissues, mouse renal tubular epithelial cells (mTECs) and HK-2 cells for further studies, including immunohistochemistry, Western blot analysis, Real-time PCR, chromatin immunoprecipitation (ChIP) and RNA sequencing, scanning electron microscopy (SEM) and Co-Immunoprecipitation (Co-IP) assay.
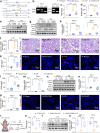
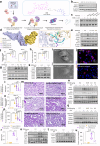
Findings: An upregulation of total Stat3 protein was observed in AKI mouse models, which correlated with patient biopsy results. This increase may be attributed to histone H3K27 acetylation. Stat3 knockout in renal tubular epithelial cells significantly reduced AKI injury and inflammation in mice. Mechanistically, Stat3 induces the transcription of tripartite motif-containing protein 21 (Trim21), triggering a cascade that activates gasdermin D (Gsdmd), resulting in pyroptosis. Administration of E034, which selectively targets Stat3 for ubiquitination and degradation, significantly alleviated renal injury in a low-dose, single-dose regimen.
Interpretation: In the context of renal injury, PROTAC emerges as a promising modality by explicitly targeting the Stat3/Trim21/Gsdmd axis, which our study has identified as a potential therapeutic target, potentially endowing clinically significant therapeutic strategies.
Funding: This work was supported by the National Key R&D Program (2022YFC2502503), the National Natural Science Foundation of China (No. 82270738), the National Natural Science Foundation of China (No. 82400806) and the Graduate Research and Practice Innovation Project of Anhui Medical University (YJS20230059).
Keywords: Histone modification; PROTAC; Pyroptosis; Renal injury; Stat3.
Copyright © 2025 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare that they have no competing interests.
Figures





References
-
- Kellum J.A., Romagnani P., Ashuntantang G., Ronco C., Zarbock A., Anders H.J. Acute kidney injury. Nat Rev Dis Primers. 2021;7(1):52. - PubMed
-
- Matsuura R., Doi K., Rabb H. Acute kidney injury and distant organ dysfunction-network system analysis. Kidney Int. 2023;103(6):1041–1055. - PubMed
-
- Wang Y., Bellomo R. Cardiac surgery-associated acute kidney injury: risk factors, pathophysiology and treatment. Nat Rev Nephrol. 2017;13(11):697–711. - PubMed
-
- Zarbock A., Nadim M.K., Pickkers P., et al. Sepsis-associated acute kidney injury: consensus report of the 28th Acute Disease Quality Initiative workgroup. Nat Rev Nephrol. 2023;19(6):401–417. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

