Expression of clock genes tracks daily and tidal time in brains of intertidal crustaceans Eurydice pulchra and Parhyale hawaiensis
- PMID: 40345195
- PMCID: PMC7617966
- DOI: 10.1016/j.cub.2025.04.047
Expression of clock genes tracks daily and tidal time in brains of intertidal crustaceans Eurydice pulchra and Parhyale hawaiensis
Abstract
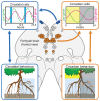
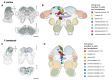
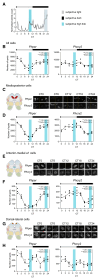
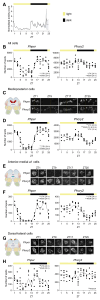
Intertidal organisms, such as the crustaceans Eurydice pulchra and Parhyale hawaiensis, express daily and tidal rhythms of physiology and behavior to adapt to their temporally complex environments. Although the molecular-genetic basis of the circadian clocks driving daily rhythms in terrestrial animals is well understood, the nature of the circatidal clocks driving tidal rhythms remains a mystery. Using in situ hybridization, we identified discrete clusters of ∼60 putative "clock" cells co-expressing canonical circadian clock genes across the protocerebrum of E. pulchra and P. hawaiensis brains. In field-collected, tidally rhythmic E. pulchra sampled under a light:dark (LD) cycle, the expression of period (per) and cryptochrome 2 (cry2) exhibited daily rhythms in particular cell groups, whereas timeless (tim) showed 12-h rhythms in others. In tidally rhythmic laboratory-reared P. hawaiensis, previously entrained to 12.4-h cycles of agitation under LD and sampled under continuous darkness, several cell groups (e.g., medioposterior cells) exhibited circadian expression of per and cry2. In contrast, dorsal-lateral cells in the protocerebrum exhibited robust ∼12-h, i.e., circatidal, rhythms of per and cry2, phased to the prior tidal agitation but not the prior LD. In P. hawaiensis exhibiting daily behavior under LD without tidal agitation, robust daily rhythms of per and cry2 expression were evident in medioposterior and other cells, whereas expression in dorsal-lateral cells was not rhythmic, underlining their essentially tidal periodicity. These results implicate canonical circadian molecules in circatidal timekeeping and reveal conserved brain networks as potential neural substrates for the generation of daily and tidal rhythms appropriate to intertidal habitats.
Keywords: Bmal1; Clock; amphipod; circadian; circatidal; cryptochrome2; isopod; period; protocerebrum; timeless.
Copyright © 2025 MRC Laboratory of Molecular Biology. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




References
-
- Dunlap JC, Loros JJ, Decoursey PJ. Chronobiology: Biological Timekeeping. Sinauer; 2004.
-
- Naylor E. Chronobiology of Marine Organisms. Cambridge University Press; 2010. - DOI
-
- Häfker NS, Andreatta G, Manzotti A, Falciatore A, Raible F, Tessmar-Raible K. Rhythms and Clocks in Marine Organisms. Ann Rev Mar Sci. 2023;15:509–538. - PubMed
-
- Enright JT. Plasticity in an isopod’s clockworks: shaking shapes form and affects phase and frequency. J Comp Physiol. 1976;107:13–37. doi: 10.1007/BF00663916. - DOI
-
- Palmer JD. Review of the dual-clock control of tidal rhythms and the hypothesis that the same clock governs both circatidal and circadian rhythms. Chronobiol Int. 1995;12:299–310. doi: 10.3109/07420529509057279. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

