Gut microbiota mediates SREBP-1c-driven hepatic lipogenesis and steatosis in response to zero-fat high-sucrose diet
- PMID: 40345386
- PMCID: PMC12145984
- DOI: 10.1016/j.molmet.2025.102162
Gut microbiota mediates SREBP-1c-driven hepatic lipogenesis and steatosis in response to zero-fat high-sucrose diet
Abstract
Objectives: Sucrose-rich diets promote hepatic de novo lipogenesis (DNL) and steatosis through interactions with the gut microbiota. However, the role of sugar-microbiota dynamics in the absence of dietary fat remains unclear. This study aimed to investigate the effects of a high-sucrose, zero-fat diet (ZFD) on hepatic steatosis and host metabolism in conventionally raised (CONVR) and germ-free (GF) mice.
Methods: CONVR and GF mice were fed a ZFD, and hepatic lipid accumulation, gene expression, and metabolite levels were analyzed. DNL activity was assessed by measuring malonyl-CoA levels, expression of key DNL enzymes, and activation of the transcription factor SREBP-1c. Metabolomic analyses of portal vein plasma identified microbiota-derived metabolites linked to hepatic steatosis. To further examine the role of SREBP-1c, its hepatic expression was knocked down using antisense oligonucleotides in CONVR ZFD-fed mice.
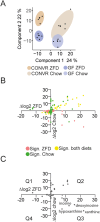
Results: The gut microbiota was essential for sucrose-induced DNL and hepatic steatosis. In CONVR ZFD-fed mice, hepatic fat accumulation increased alongside elevated expression of genes encoding DNL enzymes, higher malonyl-CoA levels, and upregulation of SREBP-1c. Regardless of microbiota status, ZFD induced fatty acid elongase and desaturase gene expression and increased hepatic monounsaturated fatty acids. Metabolomic analyses identified microbiota-derived metabolites associated with hepatic steatosis. SREBP-1c knockdown in CONVR ZFD-fed mice reduced hepatic steatosis and suppressed fatty acid synthase expression.
Conclusions: Sucrose-microbiota interactions and SREBP-1c are required for DNL and hepatic steatosis in the absence of dietary fat. These findings provide new insights into the complex interplay between diet, gut microbiota, and metabolic regulation.
Keywords: Gut microbiota; Hepatic steatosis; High-sucrose diet; Metabolomics; SREBP-1c; Zero-fat diet; de novo lipogenesis.
Copyright © 2025 The Authors. Published by Elsevier GmbH.. All rights reserved.
Conflict of interest statement
Declaration of competing interest V.T. is co-founders and shareholders of Roxbiosens Inc. M.N is co-founder and member of the Scientific Advisory Board of Caelus Pharmaceuticals and Advanced Microbial Interventions, both spin-outs of AUMC-UvA, Amsterdam, the Netherlands. F.B. is co-founder and shareholder of Roxbiosens Inc and Implexion Pharma AB, receives research funding from Biogaia AB and Novo Nordisk A/S, and is a member of the scientific advisory board of Bactolife A/S. None of these are directly relevant to the current paper.
Figures



References
-
- Le Roy T., Llopis M., Lepage P., Bruneau A., Rabot S., Bevilacqua C., et al. Intestinal microbiota determines development of non-alcoholic fatty liver disease in mice. Gut. 2013;62(12):1787–1794. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

