The role of human Shu complex in ATP-dependent regulation of RAD51 filaments during homologous recombination-associated DNA damage response
- PMID: 40345587
- PMCID: PMC12167799
- DOI: 10.1016/j.jbc.2025.110212
The role of human Shu complex in ATP-dependent regulation of RAD51 filaments during homologous recombination-associated DNA damage response
Abstract
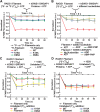
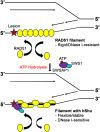
Error-free DNA lesion bypass is an important pathway in DNA damage tolerance. The Shu complex facilitates this process by promoting homologous recombination (HR) to bypass DNA damage. Biochemical analysis of the human Shu complex homolog, hSWS1-SWSAP1, offers valuable insights into the HR-associated DNA damage response. Here, we biochemically characterized the human Shu complex and examined its interactions with RAD51 filaments, which are essential in HR. Using fluorescence polarization assays, we first revealed that hSWS1-SWSAP1 preferentially binds DNA with an exposed 5' end in the presence of adenine nucleotides. We then investigated and validated the DNA-stimulated ATPase activity of hSWS1-SWSAP1 through site-specific mutagenesis, revealing that DNA with an exposed 5' end is the most efficient in enhancing this activity. Furthermore, we showed that hSWS1-SWSAP1 initially interacts with RAD51 filaments at the 5' end and modulates the properties of the nucleoprotein filaments using fluorescence-based assays. Our findings revealed that hSWS1-SWSAP1 induces conformational changes in RAD51 filaments in an ATP hydrolysis-dependent manner, while its stabilization of the filaments depends on ATP binding. This work provides mechanistic insights into the regulation of RAD51 filaments in HR-associated DNA damage tolerance.
Keywords: ATPase; DNA damage tolerance response; RAD51 filament; Shu complex; homologous recombination.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures



References
-
- Marteijn J.A., Lans H., Vermeulen W., Hoeijmakers J.H.J. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat. Rev. Mol. Cell Biol. 2014;15:465–481. - PubMed
-
- Carver A., Zhang X. Rad51 filament dynamics and its antagonistic modulators. Semin. Cell Dev. Biol. 2021;113:3–13. - PubMed
-
- San Filippo J., Sung P., Klein H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008;77:229–257. - PubMed
-
- Sung P., Klein H. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat. Rev. Mol. Cell Biol. 2006;7:739–750. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

