Demyelination Produces a Shift in the Population of Cortical Neurons That Synapse with Callosal Oligodendrocyte Progenitor Cells
- PMID: 40345851
- PMCID: PMC12177707
- DOI: 10.1523/ENEURO.0113-25.2025
Demyelination Produces a Shift in the Population of Cortical Neurons That Synapse with Callosal Oligodendrocyte Progenitor Cells
Abstract
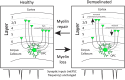
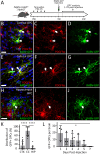
Oligodendrocyte progenitor cells (OPCs) receive synaptic input from a diverse range of neurons in the developing and adult brain. Understanding whether the neuronal populations that synapse with OPCs in the healthy brain is altered by demyelination and/or remyelination may support the advancement of neuroprotective or myelin repair strategies being developed for demyelinating diseases such as multiple sclerosis. To explore this possibility, we employed cre-lox transgenic technology to facilitate the infection of OPCs by a modified rabies virus, enabling the retrograde monosynaptic tracing of neuron→OPC connectivity. In the healthy adult mouse, OPCs in the corpus callosum primarily received synaptic input from ipsilateral cortical neurons. Of the cortical neurons, ∼50% were layer V pyramidal cells. Cuprizone demyelination reduced the total number of labeled neurons. However, the frequency/kinetics of mini-excitatory postsynaptic currents recorded from OPCs appeared preserved. Of particular interest, demyelination increased the number of labeled layer II/III pyramidal neurons and also increased at the expense of layer V pyramidal neurons, a change that was largely ameliorated by remyelination. These data suggest that in the healthy adult mouse brain, callosal OPCs primarily receive synaptic input from cortical layer V pyramidal neurons. However, callosal demyelination is associated with a population switch and OPCs equally synapse with layer II/III and V pyramidal neurons to synapse with OPCs, until myelin is restored.
Keywords: NG2 glia; demyelination; oligodendrocyte progenitor cells; rabies virus; remyelination; synapse.
Copyright © 2025 Summers et al.
Conflict of interest statement
The authors declare no competing financial interests.
Figures





Similar articles
-
C1ql1 expression in oligodendrocyte progenitor cells promotes oligodendrocyte differentiation.FEBS J. 2025 Jan;292(1):52-74. doi: 10.1111/febs.17256. Epub 2024 Sep 11. FEBS J. 2025. PMID: 39257292
-
Low-Intensity Physical Exercise is Associated with Improved Myelination and Reduced Microglial Activation in a Cuprizone-Induced Demyelination Model.Neurochem Res. 2025 Jun 5;50(3):182. doi: 10.1007/s11064-025-04441-8. Neurochem Res. 2025. PMID: 40471423 Free PMC article.
-
Single intracerebroventricular TNFR2 agonist injection impacts remyelination in the cuprizone model.J Mol Med (Berl). 2025 Jul;103(7):795-807. doi: 10.1007/s00109-025-02549-6. Epub 2025 May 10. J Mol Med (Berl). 2025. PMID: 40347238 Free PMC article.
-
The Many Lives of an Oligodendrocyte Precursor Cell.Annu Rev Neurosci. 2025 Jul;48(1):425-444. doi: 10.1146/annurev-neuro-112723-025806. Epub 2025 Apr 15. Annu Rev Neurosci. 2025. PMID: 40233142 Review.
-
High-confidence and high-throughput quantification of synapse engulfment by oligodendrocyte precursor cells.Nat Protoc. 2025 Feb;20(2):407-439. doi: 10.1038/s41596-024-01048-1. Epub 2024 Oct 3. Nat Protoc. 2025. PMID: 39363108 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
