KAT5 regulates neurodevelopmental states associated with G0-like populations in glioblastoma
- PMID: 40346033
- PMCID: PMC12064679
- DOI: 10.1038/s41467-025-59503-w
KAT5 regulates neurodevelopmental states associated with G0-like populations in glioblastoma
Abstract
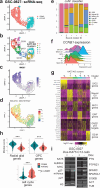
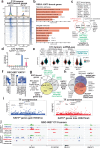
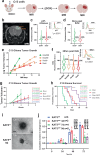
Quiescence cancer stem-like cells may play key roles in promoting tumor cell heterogeneity and recurrence for many tumors, including glioblastoma (GBM). Here we show that the protein acetyltransferase KAT5 is a key regulator of transcriptional, epigenetic, and proliferative heterogeneity impacting transitions into G0-like states in GBM. KAT5 activity suppresses the emergence of quiescent subpopulations with neurodevelopmental progenitor characteristics, while promoting GBM stem-like cell (GSC) self-renewal through coordinately regulating E2F- and MYC- transcriptional networks with protein translation. KAT5 inactivation significantly decreases tumor progression and invasive behavior while increasing survival after standard of care. Further, increasing MYC expression in human neural stem cells stimulates KAT5 activity and protein translation, as well as confers sensitivity to homoharringtonine, to similar levels to those found in GSCs and high-grade gliomas. These results suggest that the dynamic behavior of KAT5 plays key roles in G0 ingress/egress, adoption of quasi-neurodevelopmental states, and aggressive tumor growth in gliomas.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures





References
MeSH terms
Substances
Grants and funding
- R01 CA295090/CA/NCI NIH HHS/United States
- R01 CA190957/CA/NCI NIH HHS/United States
- S10 OD026919/OD/NIH HHS/United States
- R01CA190957/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- P30 CA015704/CA/NCI NIH HHS/United States
- R01NS119650/U.S. Department of Health & Human Services | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
- R35 GM139429/GM/NIGMS NIH HHS/United States
- T32CA080416/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- R21 CA232244/CA/NCI NIH HHS/United States
- R01 NS119650/NS/NINDS NIH HHS/United States
- P30CA15704/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- T32 CA080416/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

