Short-chain polyphosphates induce tau fibrillation and neurotoxicity in human iPSC-derived retinal neurons
- PMID: 40346074
- PMCID: PMC12064648
- DOI: 10.1038/s41419-025-07662-5
Short-chain polyphosphates induce tau fibrillation and neurotoxicity in human iPSC-derived retinal neurons
Abstract
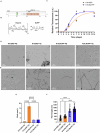
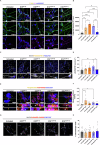
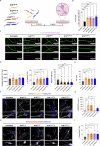
The onset of Alzheimer's Disease and Frontotemporal Dementia is closely associated with the aggregation of tau, a multifunctional protein essential for neuronal stability and function. Given the role of tau aggregation in neurodegeneration, understanding the mechanisms behind its fibril formation is crucial for developing therapeutic interventions to halt or reverse disease progression. However, the structural complexity and diverse aggregation pathways of tau present significant challenges, requiring comprehensive experimental studies. In this research, we demonstrate that short-chain polyphosphates, specifically sodium tripolyphosphate (NaTPP), effectively induce tau fibril formation in vitro using the microtubule-binding domain fragment (K18). NaTPP-induced fibrils display unique structural characteristics and aggregation kinetics compared to those induced by heparin, indicating distinct pathogenic pathways. Through molecular dynamics simulations, we show that NaTPP promotes aggregation by exposing key residues necessary for fibril formation, which remain concealed under non-aggregating conditions. This interaction drives tau into an aggregation-prone state, revealing a novel mechanism. Furthermore, our study indicates that human pluripotent stem cell-derived retinal neurons internalize NaTPP-induced fibrils within 24 h, pointing to a potential pathway for tau spread in neurodegeneration. To explore the translational implications of NaTPP-induced fibrils, we assessed their long-term effects on cellular viability, tubulin integrity, and stress responses in retinal neuron cultures. Compared to heparin, NaTPP promoted fewer but longer fibrils with initially low cytotoxicity but induced a stress response marked by increased endogenous tau and p62/SQSTM1 expression. Prolonged exposure to NaTPP-induced oligomers significantly increased cytotoxicity, leading to tubulin fragmentation, altered caspase activity, and elevated levels of phosphorylated pathological tau. These findings align with a neurodegenerative phenotype, highlighting the relevance of polyphosphates in tau pathology. Overall, this research enhances our understanding of the role of polyphosphate in tau aggregation, linking it to key cellular pathways in neurodegeneration.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The funders had no role in the study design, in the collection, analysis, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results. YG is employed by D-Tails s.r.l.; SDA is a scientific advisor of D-Tails s.r.l. The remaining authors declare that the research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest. Ethics approval and consent to participate: The use of hiPSC has been approved by the Ethical Committee for Translational Research (CERT) Sapienza University (Aut. n. 5/2022). This study does not involve animal or human participants, therefore, obtaining informed consent for participation and publication is not applicable. All methods were performed in accordance with the relevant guidelines and regulations.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

