Large-scale discovery of potent, compact and erythroid specific enhancers for gene therapy vectors
- PMID: 40346084
- PMCID: PMC12064758
- DOI: 10.1038/s41467-025-59235-x
Large-scale discovery of potent, compact and erythroid specific enhancers for gene therapy vectors
Abstract
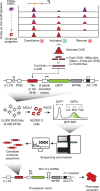
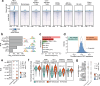
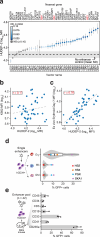
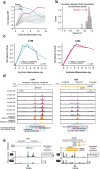
Gene expression during cell development and differentiation is orchestrated by distal regulatory elements that precisely modulate cell selective gene activity. Gene therapy vectors leverage these elements for precise spatiotemporal transgene expression. Here, we develop a one-shot approach to screen candidate regulatory sequences from large-scale epigenomics data for programmable transgene expression within gene therapy viral vectors. We assess a library of 15,000 short sequences derived from developmentally active elements during erythropoiesis using a clinically relevant reporter vector. These elements display a gradient of transcriptional enhancer activity in erythroid cells, with high cell type restriction and developmental stage specificity. Finally, replacing the canonical β-globin μLCR with a compact enhancer in a β-thalassemia lentiviral vector successfully corrects the thalassemic phenotype in patient-derived hematopoietic and stem and progenitor cells (HSPCs), while increasing viral titers and cell transducibility. Our approach provides further insights into enhancer biology with wider implications for human gene therapy.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: N.P., G.G., J.S. and J.V. have filed a patent application (US PPA No. 63224537 and PCT No. US202114752) covering various aspects of this work. The remaining authors declare no competing interests.
Figures

References
-
- Locatelli, F. et al. Betibeglogene autotemcel gene therapy for non–β0/β0 genotype β-thalassemia. N. Engl. J. Med.386, 415–427 (2021). - PubMed
-
- Kwiatkowski, J. L. et al. Betibeglogene autotemcel gene therapy in patients with transfusion-dependent, severe genotype β-thalassaemia (HGB-212): a non-randomised, multicentre, single-arm, open-label, single-dose, phase 3 trial. Lancet404, 2175–2186 (2024). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

