Profiling and functional characterization of long noncoding RNAs during human tooth development
- PMID: 40346090
- PMCID: PMC12064826
- DOI: 10.1038/s41368-025-00375-7
Profiling and functional characterization of long noncoding RNAs during human tooth development
Abstract
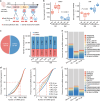
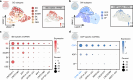
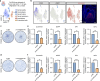
The regulatory processes in developmental biology research are significantly influenced by long non-coding RNAs (lncRNAs). However, the dynamics of lncRNA expression during human tooth development remain poorly understood. In this research, we examined the lncRNAs present in the dental epithelium (DE) and dental mesenchyme (DM) at the late bud, cap, and early bell stages of human fetal tooth development through bulk RNA sequencing. Developmental regulators co-expressed with neighboring lncRNAs were significantly enriched in odontogenesis. Specific lncRNAs expressed in the DE and DM, such as PANCR, MIR205HG, DLX6-AS1, and DNM3OS, were identified through a combination of bulk RNA sequencing and single-cell analysis. Further subcluster analysis revealed lncRNAs specifically expressed in important regions of the tooth germ, such as the inner enamel epithelium and coronal dental papilla (CDP). Functionally, we demonstrated that CDP-specific DLX6-AS1 enhanced odontoblastic differentiation in human tooth germ mesenchymal cells and dental pulp stem cells. These findings suggest that lncRNAs could serve as valuable cell markers for tooth development and potential therapeutic targets for tooth regeneration.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Ethics approval: The present study is approved by the Ethics Commission of Beijing Friendship Hospital, Capital Medical University (CMU) (2021-P2-198-02), the Beijing Laboratory of Oral Health (BLOH), CMU (Z2022SY064), and the Beijing Obstetrics and Gynecology Hospital (BOGH), CMU (2023-KY-082-01). Human fetal tissues were initially collected from BOGH and CMU, and were obtained under informed consent from donors. All human fetal tissues came from aborted fetuses for family planning purposes.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

