Epitope mapping of SARS-CoV-2 Spike protein using naturally-acquired immune responses to develop monoclonal antibodies
- PMID: 40346118
- PMCID: PMC12064773
- DOI: 10.1038/s41598-025-00555-9
Epitope mapping of SARS-CoV-2 Spike protein using naturally-acquired immune responses to develop monoclonal antibodies
Abstract
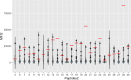
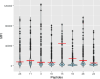
COVID-19 vaccination strategies are already available almost worldwide. However, it is also crucial to develop new therapeutic approaches, especially for vulnerable populations that may not fully respond to vaccination, such as the immunocompromised. In this project, we predicted 25 B-cell epitopes in silico in the SARS-CoV-2 Spike (S) protein and screened these against serum and plasma samples from 509 COVID-19 convalescent patients. The aim was to identify those epitopes with the highest IgG reactivity to produce monoclonal antibodies against them for COVID-19 treatment. We implemented Brewpitopes, a computational pipeline based on B-cell epitope prediction tools, such as BepiPred v2.0 and Discotope v2.0, and a series of antibody-epitope accessibility filters. We mapped the SARS-CoV-2 S protein epitopes most likely to be recognised by human neutralizing antibodies. Linear and structural epitope predictions were included and were further refined considering accessibility factors influencing their binding to antibodies like glycosylation status, localization in the viral membrane and accessibility on the 3D-surface of S. Blood samples were collected from 509 COVID-19 patients prospectively recruited 35 days after symptoms initiation, positive RT-qPCR or hospital/ICU discharge. Presence of IgG against SARS-CoV-2 was confirmed by lateral flow immunoassays. Epitopes immunogenicity was tested through the analysis of IgG levels and seropositivity in the convalescent serum and plasma samples and 126 pre-pandemic negative controls by Luminex to identify those with the highest reactivity. The seropositivity cut-offs for each epitope were calculated using a set of 126 pre-pandemic samples as negative controls (NC). Twenty-five SARS-CoV-2 S epitopes were predicted in silico as potentially the most immunogenic. These were synthesized and tested in a multiplex immunoassay against sera/plasmas from convalescent COVID-19 patients (5.7% asymptomatic, 35.6% mild, 13.8% moderate, 23% severe and 22% unknown because of anonymous donation). Among the 25 epitopes tested, 3 exhibited significantly higher IgG reactivity compared to the rest. The proportion of seropositive patients towards these 3 epitopes, based on median fluorescence intensity (MFI or Log10 MFI) above that from NC, ranged between 11 and 48%. Two out of the three most immunogenic epitopes were scaled up, resulting in the generation of two monoclonal antibodies (mAbs). These two mAbs exhibited comparable levels of S protein affinity to commercialized mAbs. Our data shows that the candidate S epitopes predicted in silico are recognised by IgG present in convalescent serum and plasma. This evidence suggests that our computational and experimental pipeline is able to yield immunogenic epitopes against SARS-CoV-2 S. These epitopes are suitable for the development of novel antibodies for preventive or therapeutic approaches against COVID-19.
Keywords: Antibody; Epitope mapping; IgG; Immunoassay; Luminex; Multiplex; Predictive models; Recovered patients of COVID-19; SARS-CoV-2; Serology; Spike protein.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Consent for publication: I, the undersigned, give my consent for the publication of identifiable details, which can include Figure(s) and/or data and/or details within the text (“Material”) to be published in the above Journal and Article. Ethics approval and consent to participate: HCB/2020/0332. Competing interests: The authors declare no competing interests.
Figures



Similar articles
-
A new multi-epitope vaccine candidate based on S and M proteins is effective in inducing humoral and cellular immune responses against SARS-CoV-2 variants: an in silico design approach.J Biomol Struct Dyn. 2024;42(22):12505-12522. doi: 10.1080/07391102.2023.2270699. Epub 2023 Oct 24. J Biomol Struct Dyn. 2024. PMID: 37874075
-
Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants.Elife. 2020 Oct 28;9:e61312. doi: 10.7554/eLife.61312. Elife. 2020. PMID: 33112236 Free PMC article.
-
Early 2022 breakthrough infection sera from India target the conserved cryptic class 5 epitope to counteract immune escape by SARS-CoV-2 variants.J Virol. 2025 Apr 15;99(4):e0005125. doi: 10.1128/jvi.00051-25. Epub 2025 Mar 26. J Virol. 2025. PMID: 40135898 Free PMC article.
-
Targeting SARS-CoV2 Spike Protein Receptor Binding Domain by Therapeutic Antibodies.Biomed Pharmacother. 2020 Oct;130:110559. doi: 10.1016/j.biopha.2020.110559. Epub 2020 Aug 1. Biomed Pharmacother. 2020. PMID: 32768882 Free PMC article. Review.
-
Structural Analysis of Neutralizing Epitopes of the SARS-CoV-2 Spike to Guide Therapy and Vaccine Design Strategies.Viruses. 2021 Jan 19;13(1):134. doi: 10.3390/v13010134. Viruses. 2021. PMID: 33477902 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
- FI19/00090/Instituto de Salud Carlos III
- 1478-2023/Sociedad Española de Neumología y Cirugía Torácica
- CB 06/06/0028/CIBER/Centro de Investigación Biomédica en Red de Enfermedades Respiratorias
- CB 06/06/0028/CIBER/Centro de Investigación Biomédica en Red de Enfermedades Respiratorias
- CB 06/06/0028/CIBER/Centro de Investigación Biomédica en Red de Enfermedades Respiratorias
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

