Deciphering the longitudinal trajectories of glioblastoma ecosystems by integrative single-cell genomics
- PMID: 40346362
- PMCID: PMC12081298
- DOI: 10.1038/s41588-025-02168-4
Deciphering the longitudinal trajectories of glioblastoma ecosystems by integrative single-cell genomics
Abstract
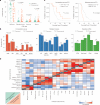
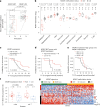
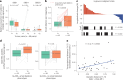
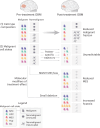
The evolution of isocitrate dehydrogenase (IDH)-wildtype glioblastoma (GBM) after standard-of-care therapy remains poorly understood. Here we analyzed matched primary and recurrent GBMs from 59 patients using single-nucleus RNA sequencing and bulk DNA sequencing, assessing the longitudinal evolution of the GBM ecosystem across layers of cellular and molecular heterogeneity. The most consistent change was a lower malignant cell fraction at recurrence and a reciprocal increase in glial and neuronal cell types in the tumor microenvironment (TME). The predominant malignant cell state differed between most matched pairs, but no states were exclusive or highly enriched in either time point, nor was there a consistent longitudinal trajectory across the cohort. Nevertheless, specific trajectories were enriched in subsets of patients. Changes in malignant state abundances mirrored changes in TME composition and baseline profiles, reflecting the co-evolution of the GBM ecosystem. Our study provides a blueprint of GBM's diverse longitudinal trajectories and highlights the treatment and TME modifiers that shape them.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: M.L.S. is an equity holder, scientific cofounder and advisory board member of Immunitas Therapeutics. I.T. is an advisory board member of Immunitas Therapeutics and a scientific cofounder of Cellyrix. R.G.W.V. is a cofounder and equity holder in Boundless Bio. The authors declare that such activities have no relationship to the present study. The other authors declare no competing interests.
Figures





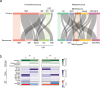
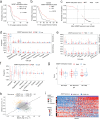

References
-
- Hegi, M. E. et al. Gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med.352, 997–1003 (2005). - PubMed
-
- Stupp, R. et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med.352, 987–996 (2005). - PubMed
-
- Mu, Q. et al. Identifying predictors of glioma evolution from longitudinal sequencing. Sci. Transl. Med.15, eadh4181 (2023). - PubMed
-
- Kim, J., Lee, I., Cho, H. J., Lee, J. & Park, P. J. Spatiotemporal evolution of the primary glioblastoma genome article spatiotemporal evolution of the primary glioblastoma genome. Cancer Cell28, 318–328 (2015). - PubMed
MeSH terms
Substances
Grants and funding
- R37 CA245523/CA/NCI NIH HHS/United States
- R35 CA253183/CA/NCI NIH HHS/United States
- R01 CA239721/CA/NCI NIH HHS/United States
- R01 CA258763/CA/NCI NIH HHS/United States
- R01 CA237208/CA/NCI NIH HHS/United States
- P30 CA013696/CA/NCI NIH HHS/United States
- P30 CA240139/CA/NCI NIH HHS/United States
- P30 CA006516/CA/NCI NIH HHS/United States
- P50 CA165962/CA/NCI NIH HHS/United States
- R21 CA256575/CA/NCI NIH HHS/United States
- R21 NS114873/NS/NINDS NIH HHS/United States
- R01 CA276765/CA/NCI NIH HHS/United States
- R01 CA280560/CA/NCI NIH HHS/United States
- P30 CA034196/CA/NCI NIH HHS/United States
- R01 CA268592/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

