Identification of programmed cell death-related genes and construction of a prognostic model in oral squamous cell carcinoma using single-cell and transcriptome analysis
- PMID: 40346375
- PMCID: PMC12064537
- DOI: 10.1007/s12672-025-02520-4
Identification of programmed cell death-related genes and construction of a prognostic model in oral squamous cell carcinoma using single-cell and transcriptome analysis
Abstract
Background: Oral squamous cell carcinoma (OSCC) is characterized by poor prognosis and high mortality. Understanding programmed cell death-related genes could provide valuable insights into disease progression and treatment strategies.
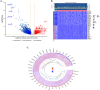
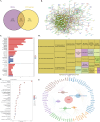
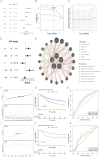
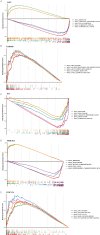
Methods: RNA-sequencing data from 341 OSCC tumor tissues and 31 healthy samples were analyzed from TCGA database, with validation using 76 samples from GSE41613. Single-cell RNA sequencing data was obtained from GSE172577 (6 OSCC samples). Differentially expressed genes (DEGs) were identified and intersected with 1,254 programmed cell death-related genes. A protein-protein interaction network was constructed, and key modules were identified. Univariate Cox, LASSO, and multivariate Cox regression analyses were performed to build a prognostic model. Model performance was evaluated using Kaplan-Meier analysis, ROC curves, and nomogram validation.
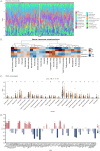
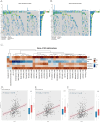
Results: The study identified 200 candidate genes from the intersection of DEGs and programmed cell death-related genes, which were further refined to 57 hub genes through PPI network analysis. A prognostic signature consisting of five genes (MET, GSDMB, KIT, PRKAG3, and CDKN2A) was established and validated. The model demonstrated good predictive performance in both training and validation cohorts (AUC > 0.6 for 1-, 2-, and 3-year survival). Single-cell analysis revealed that prognostic genes were predominantly expressed in stromal and epithelial cells. Cell communication analysis indicated strong interactions between stromal and epithelial cells.
Conclusions: This study developed and validated a novel five-gene prognostic signature for OSCC based on programmed cell death-related genes. The model shows promising clinical application potential for risk stratification and personalized treatment of OSCC patients.
Keywords: Oral squamous cell carcinoma; Prognostic model; Programmed cell death; Single-cell RNA sequencing; Tumor microenvironment.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval: Not applicable. Competing interests: The authors declare no competing interests.
Figures


References
-
- Shao S-H, Qiu S-T, Li Y, Zhang Y-Y, Cao Y-J, DI C-H, et al. Ferroptosis: mechanisms and application in tumor treatment. Zhongguo yi xue ke xue yuan xue bao Acta Academiae Medicinae Sinicae. 2024;46(5):732–9. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous
