Multi-omics integrated analysis reveals the molecular mechanism of tail fat deposition differences in sheep with different tail types
- PMID: 40346476
- PMCID: PMC12065285
- DOI: 10.1186/s12864-025-11658-y
Multi-omics integrated analysis reveals the molecular mechanism of tail fat deposition differences in sheep with different tail types
Abstract
Background: The accumulation of tail fat in sheep is a manifestation of adaptive evolution to the environment. Sheep with different tail types show significant differences in physiological functions and tail fat deposition. Although these differences reflect the developmental mechanism of tail fat under different gene regulation, the situation of sheep tail fat tissue at the single cell level has not been explored, and its molecular mechanism still needs to be further elucidated.
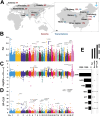
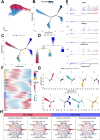
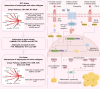
Results: Here, we characterized the genomic features of sheep with different tail types, detected the transcriptomic differences in tail adipose tissue between fat-tailed and thin-tailed sheep, established a single-cell atlas of sheep tail adipose tissue, and screened potential molecular markers (SESN1, RPRD1A and RASGEF1B) that regulate differences in sheep tail fat deposition through multi-omics integrated analysis. We found that the differential mechanism of sheep tail fat deposition not only involves adipocyte differentiation and proliferation, but is also closely related to cell-specific communication networks (When adipocytes act as signal outputters, LAMININ and other signal pathways are strongly expressed in guangling large tailed sheep and hu sheep), including interactions with immune cells and tissue remodeling to drive the typing of tail fat. In addition, we revealed the differentiation trajectory of sheep tail adipocytes through pseudo-time analysis and constructed the cell communication network of sheep tail adipose tissue.
Conclusions: Our results provide insights into the molecular mechanisms of tail fat deposition in sheep with different tail types, and provide a deeper explanation for the development and functional regulation of adipocytes.
Keywords: Molecular mechanism; Multi-omics; Sheep; Single cell; Tail fat deposition.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The experimental animals used in this study were all raised at the Animal Station of Shanxi Agricultural University. The ownership of the animals belongs to the co-author of this study (Liying Qiao), who has informed and consented to the conduct of this experiment. All experimental protocols were reviewed and approved by the Experimental Animal Ethics Committee of Shanxi Agricultural University (Ethics Committee Approval Reference Number: SXAU-EAW-2022 S.UV.010009). All authors approved the conduct of the experiment and the publication of the manuscript. In this study, in compliance with the requirements of the Animal Science and Veterinary College IACUC, sheep were humanely euthanized by exsanguination after deep anesthesia, and sodium pentobarbital (≥ 90 mg/KG) was injected intravenously to relieve the animals suffering. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures





References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

