Differences in virulence and drug resistance between Clostridioides difficile ST37 and ST1 isolates
- PMID: 40346827
- PMCID: PMC12068338
- DOI: 10.1080/21505594.2025.2502554
Differences in virulence and drug resistance between Clostridioides difficile ST37 and ST1 isolates
Abstract
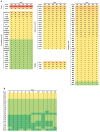
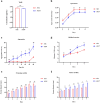
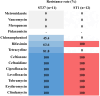
One of the most common hospital-acquired infections is caused by toxigenic Clostridioides difficile. Although C. difficile ST37 only produces a functional toxin B, it causes disease as severe as that caused by hypervirulent ST1. We aim to compare the differences in virulence and drug resistance between ST37 and ST1 isolates. We conducted whole-genome sequencing on ST37 and ST1 isolates, analyzing their type-specific genes, and the distribution and mutation of genes related to virulence and antibiotic resistance. We compared the in vitro virulence-related phenotypes of ST37 and ST1 isolates, including: TcdB concentration, number of spores formed, aggregation rate, biofilm formation, swimming diameter in semi-solid medium, motility diameter on the surface of solid medium, and their resistance to 14 CDI-related antibiotics. We detected 4 ST37-specific genes related to adherence, including lytC, cbpA, CD3246, and srtB. We detected 97 virulence-related genes in ST37 isolates that exhibit genomic differences compared to ST1. ST37 isolates showed increased aggregation, biofilm formation, and surface motility compared to ST1 in vitro. Chloramphenicol resistance gene catQ and tetracycline resistance gene tetM are present in ST37 but absent in ST1 strains. The resistance rates of ST37 to chloramphenicol and tetracycline were 45.4% and 81.8%, respectively, whereas ST1 isolates were sensitive to both antibiotics. ST1 was more resistant to rifaximin than ST37. ST37 isolates showed stronger aggregation, biofilm formation and surface motility, and had higher resistance rates to chloramphenicol and tetracycline. ST1 isolates showed stronger ability to produce toxin and sporulation, and was highly resistant to rifaximin.
Keywords: Clostridioides difficile; ST1; ST37; antibiotic resistance; virulence.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures


Similar articles
-
[Screening and phenotypic characterization of Nontoxigenic Clostridioides difficile for intervention in C. difficile infection].Zhonghua Yu Fang Yi Xue Za Zhi. 2025 Jul 6;59(7):982-988. doi: 10.3760/cma.j.cn112150-20240716-00573. Zhonghua Yu Fang Yi Xue Za Zhi. 2025. PMID: 40661004 Chinese.
-
Rifaximin resistance in Clostridioides difficile is associated with specific rpoB alleles and multilocus sequence typing (MLST) clades.BMC Microbiol. 2025 Jul 29;25(1):458. doi: 10.1186/s12866-025-04164-4. BMC Microbiol. 2025. PMID: 40731265 Free PMC article.
-
Genomic insights into tigecycline non-susceptibility in Clostridioides difficile: the role of the Tet P determinant and efflux mechanisms.BMC Microbiol. 2025 Jul 7;25(1):421. doi: 10.1186/s12866-025-04143-9. BMC Microbiol. 2025. PMID: 40624625 Free PMC article.
-
Antibiotic treatment for Clostridium difficile-associated diarrhoea in adults.Cochrane Database Syst Rev. 2017 Mar 3;3(3):CD004610. doi: 10.1002/14651858.CD004610.pub5. Cochrane Database Syst Rev. 2017. PMID: 28257555 Free PMC article.
-
Fecal microbiota transplantation for the treatment of recurrent Clostridioides difficile (Clostridium difficile).Cochrane Database Syst Rev. 2023 Apr 25;4(4):CD013871. doi: 10.1002/14651858.CD013871.pub2. Cochrane Database Syst Rev. 2023. PMID: 37096495 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- BioProject/PRJNA1160995
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
