The predictive power of profiling the DNA methylome in human health and disease
- PMID: 40346834
- PMCID: PMC12143711
- DOI: 10.1080/17501911.2025.2500907
The predictive power of profiling the DNA methylome in human health and disease
Abstract
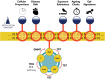
Early and accurate diagnosis significantly improves the chances of disease survival. DNA methylation (5mC), the major DNA modification in the human genome, is now recognized as a biomarker of immense clinical potential. This is due to its ability to delineate precisely cell-type, quantitate both internal and external exposures, as well as tracking chronological and biological components of the aging process. Here, we survey the current state of DNA methylation as a biomarker and predictor of traits and disease. This includes Epigenome-wide association study (EWAS) findings that inform Methylation Risk Scores (MRS), EpiScore long-term estimators of plasma protein levels, and machine learning (ML) derived DNA methylation clocks. These all highlight the significant benefits of accessible peripheral blood DNA methylation as a surrogate measure. However, detailed DNA methylation biopsy analysis in real-time is also empowering pathological diagnosis. Furthermore, moving forward, in this multi-omic and biobank scale era, novel insights will be enabled by the amplified power of increasing sample sizes and data integration.
Keywords: DNA methylation; EWAS; Epigenetics; biological ageing; epigenetic clocks; epigenetic estimators; epigenomics; methylation risk scores.
Conflict of interest statement
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties. No writing assistance was utilized in the production of this manuscript.
No writing assistance was utilized in the production of this manuscript.
Figures
References
-
- Hillary RF, Ng HK, McCartney DL, et al. Blood-based epigenome-wide analyses of chronic low-grade inflammation across diverse population cohorts. Cell Genom. 2024;4(5):100544. doi: 10.1016/j.xgen.2024.100544 - DOI - PMC - PubMed
-
•• Illustrates the potential advantage of EpiScores in capturing long term trends.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
