Exploiting host kinases to combat dengue virus infection and disease
- PMID: 40348023
- PMCID: PMC12245208
- DOI: 10.1016/j.antiviral.2025.106172
Exploiting host kinases to combat dengue virus infection and disease
Abstract
The burden of dengue on human health has dramatically increased in recent years, underscoring the urgent need for effective therapeutic interventions. Despite decades of research since the discovery of the dengue virus, no specific antiviral treatments are available and strategies to reliably prevent severe disease remain limited. Direct-acting antivirals against dengue are under active investigation but have shown limited efficacy to date. An underappreciated Achille's heal of the virus is its dependence on host factors for infection and pathogenesis, each of which presents a potential avenue for therapeutic intervention. We and others have demonstrated that dengue virus relies on multiple host kinases, some of which are already targeted by clinically approved inhibitors. These offer drug repurposing opportunities for host-directed dengue treatment. Here, we summarize findings on the role of kinases in dengue infection and disease and highlight potential kinase targets for the development of innovative host-directed therapeutics.
Keywords: Dengue virus; Flavivirus; Host-targeted antivirals; Kinase inhibitors; Kinase signaling; Neglected tropical disease.
Copyright © 2025 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


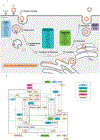
References
-
- WHO TEAM: Immunization, V. a. B. I (World Health Organization, 2024).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

