The bacterial chaperone CsgC inhibits functional amyloid CsgA formation by promoting the intrinsically disordered pre-nuclear state
- PMID: 40348191
- PMCID: PMC12205649
- DOI: 10.1016/j.jbc.2025.110217
The bacterial chaperone CsgC inhibits functional amyloid CsgA formation by promoting the intrinsically disordered pre-nuclear state
Abstract
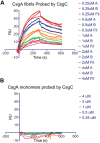
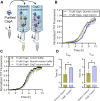
Escherichia coli assembles a functional amyloid called curli during biofilm formation. The major curlin subunit is the CsgA protein, which adopts a beta-sheet-rich fold upon fibrillization. The chaperone protein CsgC inhibits CsgA amyloid formation. CsgA undergoes a 3-stage aggregation process: an initial lag phase where beta-rich nuclei form, an exponential elongation phase, and a plateau phase. It is currently not known whether CsgC inhibits amyloid formation by inhibiting the formation of a pre-fibril nucleus or whether CsgC inhibits a later stage of amyloid formation by blocking monomer addition. Here, CsgC homologs from Citrobacter youngae, Cedecea davisae, and Hafnia alvei were purified and characterized for their ability to interrogate CsgA amyloid formation. Each of the CsgC homologs prolonged the lag phase of E. coli CsgA amyloid formation, similar to E. coli CsgC. Additionally, we found E. coli CsgC interacted transiently and weakly with a monomeric, pre-nucleus species of CsgA, which delayed amyloid formation. A transient CsgC-CsgA heterodimer was observed using ion mobility-mass spectrometry. When CsgC was added to actively polymerizing CsgA, exponential growth commonly associated with nucleation-dependent amyloid formation was lost. Adding preformed CsgA seeds did not rescue exponential growth, indicating that CsgC also has inhibitory activity during fibril elongation. Indeed, CsgC interacted strongly with CsgA fibers, suggesting that the interaction between CsgC and CsgA fibers can slow new fiber growth. CsgC displays unique inhibitory activity at multiple stages of amyloid formation and acts as an energy-independent chaperone that transiently interacts with prefibrillar CsgA and an amyloid fiber.
Keywords: amyloid; chaperone; inhibition mechanism; mass spectrometry; surface plasmon resonance.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures




Update of
-
The bacterial chaperone CsgC inhibits functional amyloid CsgA formation by promoting the intrinsically disordered pre-nuclear state.bioRxiv [Preprint]. 2025 Apr 2:2025.03.21.644623. doi: 10.1101/2025.03.21.644623. bioRxiv. 2025. Update in: J Biol Chem. 2025 Jun;301(6):110217. doi: 10.1016/j.jbc.2025.110217. PMID: 40166156 Free PMC article. Updated. Preprint.
References
-
- Chiti F., Dobson C.M. Protein misfolding, amyloid formation, and human disease: a summary of progress over the last decade. Annu. Rev. Biochem. 2017;86:27–68. - PubMed
-
- Chiti F., Dobson C.M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006;75:333–366. - PubMed
-
- Bucciantini M., Giannoni E., Chiti F., Baroni F., Taddei N., Ramponi G., et al. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nat. 2002;6880:507–511. - PubMed
-
- Haass C., Selkoe D.J. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer’s amyloid β-peptide. Nat. Rev. Mol. Cell Biol. 2007;2:101–112. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

