An approach to characterize mechanisms of action of anti-amyloidogenic compounds in vitro and in situ
- PMID: 40348747
- PMCID: PMC12065871
- DOI: 10.1038/s41531-025-00966-5
An approach to characterize mechanisms of action of anti-amyloidogenic compounds in vitro and in situ
Abstract
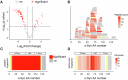
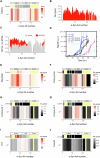
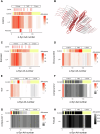
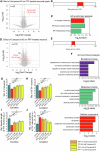
Amyloid aggregation is associated with neurodegenerative disease and its modulation is a focus of drug development. We developed a chemical proteomics pipeline to probe the mechanism of action of anti-amyloidogenic compounds. Our approach identifies putative interaction sites with high resolution, can probe compound interactions with specific target conformations and directly in cell and brain extracts, and identifies off-targets. We analysed interactions of six anti-amyloidogenic compounds and the amyloid binder Thioflavin T with different conformations of the Parkinson's disease protein α-Synuclein and tested specific compounds in cell or brain lysates. AC Immune compound 2 interacted with α-Synuclein in vitro, in intact neurons and in neuronal lysates, reduced neuronal α-Synuclein levels in a seeded model, and had protective effects. EGCG, Baicalein, ThT and doxycycline interacted with α-Synuclein in vitro but not substantially in cell lysates, with many additional putative targets, underscoring the importance of testing compounds in situ. Our pipeline will enable screening of compounds against any amyloidogenic proteins in cell and patient brain extracts and mechanistic studies of compound action.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: AC Immune (ACI) provided two of the compounds used in the study and Ait-Bouziad, N., Davranche, A., Boudou, C., Tsika, E., Ouared, A., and Stöhr, J. are employees of ACI. LiP-MS is subject of a patent licensed to the company Biognosys. The other authors declare no competing interests.
Figures



References
LinkOut - more resources
Full Text Sources

