Phenotypic drug susceptibility testing for Mycobacterium tuberculosis variant bovis BCG in 12 hours
- PMID: 40348759
- PMCID: PMC12065818
- DOI: 10.1038/s41467-025-59736-9
Phenotypic drug susceptibility testing for Mycobacterium tuberculosis variant bovis BCG in 12 hours
Abstract
Drug-resistant tuberculosis (DR-TB) kills ~200,000 people every year. A contributing factor is the slow turnaround time (TAT) associated with drug susceptibility diagnostics. The prevailing gold standard for phenotypic drug susceptibility testing (pDST) takes at least two weeks. Here we show that growth-based pDST for slow-growing mycobacteria can be conducted in 12 h. We use Mycobacterium tuberculosis variant bovis Bacillus Calmette-Guérin (BCG) and Mycobacterium smegmatis as the mycobacterial pathogen models and expose them to antibiotics used in (multidrug-resistant) tuberculosis (TB) treatment regimens - i.e., rifampicin (RIF), isoniazid (INH), ethambutol (EMB), linezolid (LZD), streptomycin (STR), bedaquiline (BDQ), and levofloxacin (LFX). The bacterial growth in a microfluidic chip is tracked by time-lapse phase-contrast microscopy. A deep neural network-based segmentation algorithm is used to quantify the growth rate and to determine how the strains responded to drug treatments. Most importantly, a panel of susceptible and resistant M. bovis BCG are tested at critical concentrations for INH, RIF, STR, and LFX. The susceptible strains could be identified in less than 12 h. These findings are comparable to what we expect for pathogenic M. tuberculosis as they share 99.96% genetic identity.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: J.E. has patented the method (US10,041,104B) and founded Astrego Diagnostics, but he has no current association with that company. No current company is associated with this work, but there may be in the future. All other authors declare no competing interests.
Figures


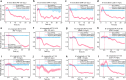
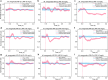

References
-
- Geneva: World Health Organization. Global Tuberculosis Report. https://www.who.int/teams/global-tuberculosis-programme/tb-reports/globa... (2024).
-
- Pedersen, O. S. et al. Global treatment outcomes of extensively drug-resistant tuberculosis in adults: a systematic review and meta-analysis. J. Infect.87, 177–189 (2023). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

