Anti-Müllerian hormone signalling sustains circadian homeostasis in zebrafish
- PMID: 40348785
- PMCID: PMC12065890
- DOI: 10.1038/s41467-025-59528-1
Anti-Müllerian hormone signalling sustains circadian homeostasis in zebrafish
Abstract
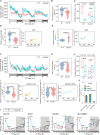
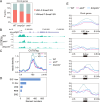
Circadian clocks temporally orchestrate the behavioural and physiological rhythms. The core molecules establishing the circadian clock are clear; however, the critical signalling pathways that cause or favour the homeostasis are poorly understood. Here, we report that anti-Müllerian hormone (Amh)-mediated signalling plays an important role in sustaining circadian homeostasis in zebrafish. Notably, amh knockout dampens molecular clock oscillations and disrupts both behavioural and hormonal circadian rhythms, which are recapitulated in bmpr2a null mutants. Somatotropes and gonadotropes are identified as Amh-targeted pituitary cell populations. Single-cell transcriptome analysis further reveals a lineage-specific regulation of pituitary clock by Amh. Moreover, Amh-induced effect on clock gene expression can be abolished by blocking Smad1/5/9 phosphorylation and bmpr2a knockout. Mechanistically, Amh binds to its receptors, Bmpr2a/Bmpr1bb, which in turn activate Smad1/5/9 by phosphorylation and promote circadian gene expression. Our findings reveal a key hormone signalling pathway for circadian homeostasis in zebrafish with implications for rhythmic organ functions and circadian health.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures





Similar articles
-
Genetic evidence for Amh modulation of gonadotropin actions to control gonadal homeostasis and gametogenesis in zebrafish and its noncanonical signaling through Bmpr2a receptor.Development. 2020 Nov 29;147(22):dev189811. doi: 10.1242/dev.189811. Development. 2020. PMID: 33060133
-
Casein kinase 1δ activity: a key element in the zebrafish circadian timing system.PLoS One. 2013;8(1):e54189. doi: 10.1371/journal.pone.0054189. Epub 2013 Jan 21. PLoS One. 2013. PMID: 23349822 Free PMC article.
-
Disrupting Amh and androgen signaling reveals their distinct roles in zebrafish gonadal differentiation and gametogenesis.Commun Biol. 2025 Mar 5;8(1):371. doi: 10.1038/s42003-025-07719-3. Commun Biol. 2025. PMID: 40044757 Free PMC article.
-
Coupling the Circadian Clock to Homeostasis: The Role of Period in Timing Physiology.Endocr Rev. 2019 Feb 1;40(1):66-95. doi: 10.1210/er.2018-00049. Endocr Rev. 2019. PMID: 30169559 Review.
-
Role of core circadian clock genes in hormone release and target tissue sensitivity in the reproductive axis.Mol Cell Endocrinol. 2020 Feb 5;501:110655. doi: 10.1016/j.mce.2019.110655. Epub 2019 Nov 19. Mol Cell Endocrinol. 2020. PMID: 31756424 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

