Recommendations for sample selection, collection and preparation for NMR-based metabolomics studies of blood
- PMID: 40348843
- PMCID: PMC12065766
- DOI: 10.1007/s11306-025-02259-7
Recommendations for sample selection, collection and preparation for NMR-based metabolomics studies of blood
Abstract
Background: Metabolic profiling of blood metabolites, particularly in plasma and serum, is vital for studying human diseases, human conditions, drug interventions and toxicology. The clinical significance of blood arises from its close ties to all human cells and facile accessibility. However, patient-specific variables such as age, sex, diet, lifestyle and health status, along with pre-analytical conditions (sample handling, storage, etc.), can significantly affect metabolomic measurements in whole blood, plasma, or serum studies. These factors, referred to as confounders, must be mitigated to reveal genuine metabolic changes due to illness or intervention onset.
Review objective: This review aims to aid metabolomics researchers in collecting reliable, standardized datasets for NMR-based blood (whole/serum/plasma) metabolomics. The goal is to reduce the impact of confounding factors and enhance inter-laboratory comparability, enabling more meaningful outcomes in metabolomics studies.
Key concepts: This review outlines the main factors affecting blood metabolite levels and offers practical suggestions for what to measure and expect, how to mitigate confounding factors, how to properly prepare, handle and store blood, plasma and serum biosamples and how to report data in targeted NMR-based metabolomics studies of blood, plasma and serum.
Keywords: Blood; Metabolites; Metabolomics; NMR; Plasma; Serum; Standardization.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Conflict of interest: Dr. Kaddurah-Daouk is an inventor on a series of patents on use of metabolomics for the diagnosis and treatment of CNS diseases and holds equity in Metabolon Inc., Chymia LLC and PsyProtix.
Figures

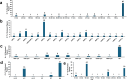
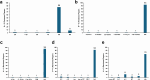
References
-
- Abdul Rahim, M. B. H., Chilloux, J., Martinez-Gili, L., Neves, A. L., Myridakis, A., Gooderham, N., & Dumas, M. E. (2019). Diet-induced metabolic changes of the human gut microbiome: Importance of short-chain fatty acids, methylamines and indoles. Acta Diabetologica,56(5), 493–500. 10.1007/s00592-019-01312-x - PMC - PubMed
-
- Ala-Korpela, M., Hiltunen, Y., & Bell, J. D. (1995). Quantification of biomedical NMR data using artificial neural network analysis: Lipoprotein lipid profiles from 1H NMR data of human plasma. NMR in Biomedicine, 8(6), 235–244. 10.1002/nbm.1940080603 - PubMed
-
- Amathieu, R., Triba, M. N., Goossens, C., Bouchemal, N., Nahon, P., Savarin, P., & Moyec, L. L. (2016). Nuclear magnetic resonance based metabolomics and liver diseases: Recent advances and future clinical applications. World Journal of Gastroenterology,22(1), 417–426. 10.3748/wjg.v22.i1.417 - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
