Temperature-induced variation in the transcriptome of maritime pine (Pinus pinaster Ait.) embryogenic masses modulates the phenotype of the derived plants
- PMID: 40348991
- PMCID: PMC12065292
- DOI: 10.1186/s12864-025-11610-0
Temperature-induced variation in the transcriptome of maritime pine (Pinus pinaster Ait.) embryogenic masses modulates the phenotype of the derived plants
Abstract
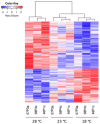
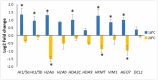
A number of studies show that combining somatic embryogenesis with environmental stimuli can induce plant defenses against abiotic stresses, offering a complementary strategy in tree breeding programs. In a previous study, we found that increasing/decreasing the standard temperature of 23 ˚C by 5 ˚C during maritime pine (Pinus pinaster) somatic embryo maturation resulted in epitypes, as the derived plants showed altered phenotypes regarding leaf histology, proline content, photosynthetic rates, and hormone profiles, and that also differentially respond after a short-term heat stress. To elucidate the mechanisms underlying these altered phenotypes, we sequenced the transcriptome of embryonal-suspensor masses (EMs) from the three epitypes, identifying 812 differentially expressed genes (DEGs). Ten genes involved in epigenetic regulation were specifically up-regulated in EMs of the cold epitype. While some of these genes have been linked with somatic embryo maturation, the increased expression of three of these genes, histone deacetylases HDA9, a histone-lysine methyl-transferase (HKMT) and an Argonaute (AGO7), was found to be low temperature-induced epigenetic marks. Among the genes up-regulated in the EMs from the warm epitype, we studied those related to abiotic stress response and observed greater variation in genes involved in abscisic acid (ABA)-mediated response such as those encoding Ras GTPase-activating protein-binding (G3BP) proteins, an AAA-ATPase, and an aspartyl protease (APF2). We also found differential expression in genes encoding for RING-type E3 ubiquitin-transferases, and DNAJ and BAG chaperones. Additionally, the biosynthetic pathways of jasmonic acid, cytokinins and the diterpene pimaradiene were also altered in the warm epitype. However, the increased ABA and cytokinin content observed in the plants derived from this warm epitype cannot be fully explained by the EMs transcriptome profile. Conversely, in the cold epitype, we observed downregulation of genes encoding for an ABA receptor (PYL3), and a xyloglucan endotrans-glucosylase/hydrolase (XTH6). These findings support the hypothesis that the previously reported heat-adapted phenotype of plants derived from the cold epitype (characterized by a faster and higher proline increase, lower increases in ABA levels, no reduction in active cytokinins, and a better net photosynthesis rate recovery) could be attributed to low-temperature-induced epigenetic marks that were absent in the warm epitype.
Keywords: Climate change; Epitypes; Forest-trees; Gene expression; RNA-seq.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures



References
-
- Maes J, Teller A, Erhard M, Condé S, Vallecillo S, Barredo Cano JI, Paracchini M, Abdul Malak D, Trombetti M, Vigiak O, Zulian G, Addamo A, Grizzetti B, Somma F, Hagyo A, Vogt P, Polce C, Jones A, Marin A, Ivits E, Mauri A, Rega C, Czucz B, Ceccherini G, Pisoni E, Ceglar A, De Palma P, Cerrani I, Meroni M, Caudullo G, Lugato E, Vogt J, Spinoni J, Cammalleri C, Bastrup-Birk A, San-Miguel-Ayanz J, San Román S, Kristensen P, Christiansen T, Zal N, De Roo A, De Jesus Cardoso A, Pistocchi A, Del Barrio Alvarellos I, Tsiamis K, Gervasini E, Deriu I, La Notte A, Viñas A, Vizzarri R, Camia M, Robert A, Kakoulaki N, Garcia Bendito G, Panagos E, Ballabio P, Scarpa C, Montanarella S, Orgiazzi L. A., Fernandez Ugalde, O. and Santos-Martín, F. 2020. Mapping and Assessment of Ecosystems and their Services: An EU ecosystem assessment. Joint Research Centre Policy Report, European Commission. 10.2760/757183
-
- Senf C, Sebald J, Seidl R. Increasing canopy mortality affects the future demographic structure of Europe’s forests. One Earth. 2021;4:749–55. 10.1016/j.oneear.2021.04.008.
-
- Giorgi F, Lionello P. Climate change projections for the mediterranean region. Glob Planet Chang. 2008;63:90–104. 10.1016/j. - DOI
-
- Yakovlev I, Fossdal CG, Skrøppa T, Olsen JE, Jahren AH, Johnsen Ø. An adaptive epigenetic memory in conifers with important implications for seed production. Seed Sci Res. 2012;22:63–76. 10.1017/S0960258511000535.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

