Development of a DNA Aptamer-Based Approach to Noninvasively Image CAR-T Cells In Vivo and Traceless Enrichment In Vitro
- PMID: 40349166
- PMCID: PMC12302554
- DOI: 10.1002/advs.202506746
Development of a DNA Aptamer-Based Approach to Noninvasively Image CAR-T Cells In Vivo and Traceless Enrichment In Vitro
Abstract
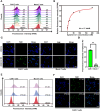
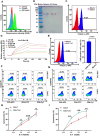
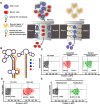
Chimeric antigen receptor (CAR) T cells offered a potential cure for malignancies, however, their outcomes and dynamics across different anatomical sites remained inadequately characterized. Monitoring the bio-distribution and tumor-homing of CAR-T cells in vivo is crucial, as it provides patient-specific data that might inform on treatment success, potential failure, and off-target toxicities. Herein, an Aptamer A3 by Cell-SELEX (systematic evolution of ligands by exponential enrichment) is generated, which can bind with CAR-T cells with nanomolar affinity. After CAR-T cells are injected into Nalm6 xenograft tumor model mice through tail vein, Cy5-labeled A3 is injected into mice for fluorescence time-delay imaging in vivo. The fluorescence signal produced by the Cy5-labeled A3 is accumulated in the tumor area and reached its maximum at day 14. Moreover, A3 could enrich CAR-T cells in mixed cell populations in a traceless way. A3 is screened for CAR-T cells imaging and CAR-T cells enrichment, which may provide assistance for the evaluation of CAR-T cells efficacy and the manufacture of CAR-T cells. Overall, this research shows that A3 enabled repeated, sensitive, and specific assessment of the infused CAR-T cells in vivo. The screened aptamer will have broad applications for tracking CAR-T cells in patients, providing insights into treatment success, potential failure, and off-target toxicities.
Keywords: CAR‐T cells; DNA aptamers; enrichment; fluorescence imaging; tracking.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
CAR-T Cells Therapy in Glioblastoma: A Systematic Review on Molecular Targets and Treatment Strategies.Int J Mol Sci. 2024 Jun 29;25(13):7174. doi: 10.3390/ijms25137174. Int J Mol Sci. 2024. PMID: 39000281 Free PMC article.
-
A new sort of cells for chimeric antigen receptor T-cell therapies-isolating CD14-CD127+ T cells for chimeric antigen receptor T-cell manufacture.Cytotherapy. 2025 Aug;27(8):980-990. doi: 10.1016/j.jcyt.2025.04.068. Epub 2025 Apr 18. Cytotherapy. 2025. PMID: 40380956
-
Chimeric antigen receptor (CAR) T-cell therapy for people with relapsed or refractory diffuse large B-cell lymphoma.Cochrane Database Syst Rev. 2021 Sep 13;9(9):CD013365. doi: 10.1002/14651858.CD013365.pub2. Cochrane Database Syst Rev. 2021. PMID: 34515338 Free PMC article.
-
In vitro machine learning-based CAR T immunological synapse quality measurements correlate with patient clinical outcomes.PLoS Comput Biol. 2022 Mar 18;18(3):e1009883. doi: 10.1371/journal.pcbi.1009883. eCollection 2022 Mar. PLoS Comput Biol. 2022. PMID: 35303007 Free PMC article. Clinical Trial.
-
Placental circulating T cells: a novel, allogeneic CAR-T cell platform with preserved T-cell stemness, more favorable cytokine profile, and durable efficacy compared to adult PBMC-derived CAR-T.J Immunother Cancer. 2024 Apr 29;12(4):e008656. doi: 10.1136/jitc-2023-008656. J Immunother Cancer. 2024. PMID: 38684370 Free PMC article.
References
-
- Davila M. L., Riviere I., Wang X., Bartido S., Park J., Curran K., Chung S. S., Stefanski J., Borquez‐Ojeda O., Olszewska M., Qu J., Wasielewska T., He Q., Fink M., Shinglot H., Youssif M., Satter M., Wang Y., Hosey J., Quintanilla H., Halton E., Bernal Y., Bouhassira D. C., Arcila M. E., Gonen M., Roboz G. J., Maslak P., Douer D., Frattini M. G., Giralt S., et al., Sci. Transl. Med. 2014, 6, 224ra25. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
