UCHL1 regulates adiponectin receptors in Sertoli cells to maintain testicular homeostatic balance
- PMID: 40349775
- PMCID: PMC12167481
- DOI: 10.1016/j.jbc.2025.110221
UCHL1 regulates adiponectin receptors in Sertoli cells to maintain testicular homeostatic balance
Abstract
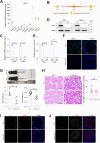
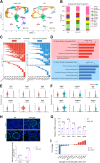
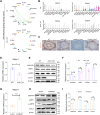
Disruptions in testicular homeostasis can lead to impaired spermatogenesis and male infertility. Such disturbances may result from various factors, including viral or bacterial infections, toxic injuries, and genetic mutations or deletions. The maintenance of testicular homeostasis is governed by a complex interplay of various cells, hormones, paracrine factors, genes, and enzymes. UCHL1, a member of the deubiquitinating enzyme family, is recognized for its role in neuronal function. However, its contribution to testicular homeostasis and spermatogenesis remains unclear. This study uncovers a critical role for Uchl1 in maintaining testicular homeostasis, acting as a regulatory switch for spermatogenesis. We demonstrate that Uchl1 knockout (Uchl1_KO) mice exhibit reduced body weight, decreased testicular specific gravity, and impaired spermatogenesis. Single-nucleus RNA sequencing (snRNA-seq) analysis of Uchl1_KO testes reveals a significant decrease in oxidative phosphorylation (OXPHOS) levels and an increase in Sertoli cell abnormalities. Notably, Uchl1_KO/knockdown downregulates metabolism-related adiponectin signaling (ADIPOR1/AMPK) and upregulates the inflammation-related SEMA7A/PLXNC1 pathway. Sertoli cell lines (oeAdipor1/shUchl1) confirm UCHL1's dual regulatory role in these signaling pathways in vitro experiments. Our findings identify UCHL1 as a key regulator of testicular homeostasis and spermatogenesis, and it dynamically controls the balance between metabolic and inflammatory signaling in the testis. This study provides a valuable theoretical foundation for exploring the molecular mechanisms underlying testicular homeostasis balance and for advancing human reproductive health.
Keywords: ADIPONECTIN; SEMA7A; Uchl1; homeostasis; snRNA-seq; spermatogenesis.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

