Genomic insights into novel extremotolerant bacteria isolated from the NASA Phoenix mission spacecraft assembly cleanrooms
- PMID: 40350519
- PMCID: PMC12067966
- DOI: 10.1186/s40168-025-02082-1
Genomic insights into novel extremotolerant bacteria isolated from the NASA Phoenix mission spacecraft assembly cleanrooms
Erratum in
-
Correction: Genomic insights into novel extremotolerant bacteria isolated from the NASA Phoenix mission spacecraft assembly cleanrooms.Microbiome. 2025 Oct 21;13(1):211. doi: 10.1186/s40168-025-02274-9. Microbiome. 2025. PMID: 41121292 Free PMC article. No abstract available.
Abstract
Background: Human-designed oligotrophic environments, such as cleanrooms, harbor unique microbial communities shaped by selective pressures like temperature, humidity, nutrient availability, cleaning reagents, and radiation. Maintaining the biological cleanliness of NASA's mission-associated cleanrooms, where spacecraft are assembled and tested, is critical for planetary protection. Even with stringent controls such as regulated airflow, temperature management, and rigorous cleaning, resilient microorganisms can persist in these environments, posing potential risks for space missions.
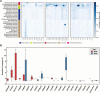
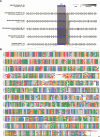
Results: During the Phoenix spacecraft mission, genomes of 215 bacterial isolates were sequenced and based on overall genome-related indices, 53 strains belonging to 26 novel species were recognized. Metagenome mapping indicated less than 0.1% of the reads associated with novel species, suggesting their rarity. Genes responsible for biofilm formation, such as BolA (COG0271) and CvpA (COG1286), were predominantly found in proteobacterial members but were absent in other non-spore-forming and spore-forming species. YqgA (COG1811) was detected in most spore-forming members but was absent in Paenibacillus and non-spore-forming species. Cell fate regulators, COG1774 (YaaT), COG3679 (YlbF, YheA/YmcA), and COG4550 (YmcA, YheA/YmcA), controlling sporulation, competence, and biofilm development processes, were observed in all spore-formers but were missing in non-spore-forming species. COG analyses further revealed resistance-conferring proteins in all spore-formers (n = 13 species) and eight actinobacterial species, responsible for enhanced membrane transport and signaling under radiation (COG3253), transcription regulation under radiation stress (COG1108), and DNA repair and stress responses (COG2318). Additional functional analysis revealed that Agrococcus phoenicis, Microbacterium canaveralium, and Microbacterium jpeli contained biosynthetic gene clusters (BGCs) for ε-poly-L-lysine, beneficial in food preservation and biomedical applications. Two novel Sphingomonas species exhibited for zeaxanthin, an antioxidant beneficial for eye health. Paenibacillus canaveralius harbored genes for bacillibactin, crucial for iron acquisition. Georgenia phoenicis had BGCs for alkylresorcinols, compounds with antimicrobial and anticancer properties used in food preservation and pharmaceuticals.
Conclusion: Despite stringent decontamination and controlled environmental conditions, cleanrooms harbor unique bacterial species that form biofilms, resist various stressors, and produce valuable biotechnological compounds. The reduced microbial competition in these environments enhances the discovery of novel microbial diversity, contributing to the mitigation of microbial contamination and fostering biotechnological innovation. Video Abstract.
Keywords: BGCs; Cleanroom; Extreme environment; Novel species; Phoenix mission; Phylogenomics.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Since the study did not involve human subjects, ethics approval and consent to participate were not applicable. The authors declare that the research was conducted without any commercial or financial relationships that could be perceived as potential conflicts of interest. Competing interests: The authors declare no competing interests.
Figures




References
-
- Mora M, Mahnert A, Koskinen K, Pausan MR, Oberauner-Wappis L, Krause R, et al. Microorganisms in confined habitats: microbial monitoring and control of intensive care units, operating rooms, cleanrooms and the international space station. Front Microbiol. 2016;7(1573). 10.3389/fmicb.2016.01573. - PMC - PubMed
-
- Miliotis G, Sengupta P, Hameed A, Chuvochina M, McDonagh F, Simpson AC, et al. Novel spore-forming species exhibiting intrinsic resistance to third- and fourth-generation cephalosporins and description of Tigheibacillus jepli gen. nov., sp. nov. mBio. 2024;15(4):e00181-24. 10.1128/mbio.00181-24. - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

