Reducing HuD Levels Alleviates Alzheimer's Disease Pathology in 5xFAD Mice
- PMID: 40351099
- PMCID: PMC12151878
- DOI: 10.1111/acel.70080
Reducing HuD Levels Alleviates Alzheimer's Disease Pathology in 5xFAD Mice
Abstract
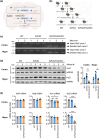
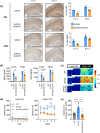
Alzheimer's disease (AD) is the most common neurodegenerative pathology in older persons. The accumulation of amyloid β (Aβ) plaques is a major contributor to AD development. The RNA-binding protein HuD/ELAVL4 has been implicated in the formation of Aβ plaques, but its role in AD is unclear. Here, we report that ablation of HuD from CAMK2A+ neurons (HuDcKO) in the 5xFAD mouse model of AD results in a significant reduction of Aβ plaques and the alleviation of some AD-associated behaviors. Given the lack of effective therapies for AD, we propose that reducing HuD levels or function can contribute to diminishing Aβ plaque formation and AD-associated pathology.
Keywords: Elavl4; 5xFAD; Alzheimer's disease; HuD; aβ plaques; homecage activity.
Published 2025. This article is a U.S. Government work and is in the public domain in the USA. Aging Cell published by Anatomical Society and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

