A label-free nanowell-based impedance sensor for ten-minute SARS-CoV-2 detection
- PMID: 40352677
- PMCID: PMC12056702
- DOI: 10.1039/d5sd00002e
A label-free nanowell-based impedance sensor for ten-minute SARS-CoV-2 detection
Abstract
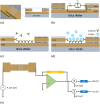
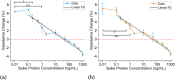
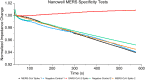
This work explores label-free biosensing as an effective method for biomolecular analysis, ensuring the preservation of native conformation and biological activity. The focus is on a novel electronic biosensing platform utilizing micro-fabricated nanowell-based impedance sensors, offering rapid, point-of-care diagnosis for SARS-CoV-2 (COVID-19) detection. The nanowell sensor, constructed on a silica substrate through a series of microfabrication processes including deposition, patterning, and etching, features a 5 × 5 well array functionalized with antibodies. Real-time impedance changes within the nanowell array enable diagnostic results within ten minutes using small sample volumes (<5 μL). The research includes assays for SARS-CoV-2 spike proteins in phosphate-buffered saline (PBS) and artificial saliva buffers to mimic real human SARS-CoV-2 samples, covering a wide range of concentrations. The sensor exhibits a detection limit of 0.2 ng mL-1 (1.5 pM) for spike proteins. Middle East respiratory syndrome (MERS-CoV) spike proteins are differentiated from SARS-CoV-2 spike proteins, demonstrating specificity.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures



References
-
- WHO COVID-19 dashboard, https://data.who.int/dashboards/covid19/cases?n=c
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
