PseudoCell: A Multivalued Logical Regulatory Network to Investigate Premature Senescence Dynamics and Heterogeneity
- PMID: 40353054
- PMCID: PMC12064993
- DOI: 10.1002/agm2.70020
PseudoCell: A Multivalued Logical Regulatory Network to Investigate Premature Senescence Dynamics and Heterogeneity
Abstract
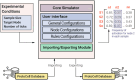
Purpose: Premature cellular senescence is a pivotal process in aging and age-related diseases, triggered by various stressors. However, this is not a homogeneous phenotype, but a heterogeneous cellular state composed of multiple senescence programs with different compositions. Therefore, understanding the complex dynamics of senescence programs requires a systemic approach. We introduce PseudoCell, a multivalued logical regulatory network designed to explore the molecular intricacies of premature senescence.
Methods: PseudoCell integrates key senescence signaling pathways and molecular mechanisms, offering a versatile platform for investigating diverse premature senescence programs initiated by different stimuli.
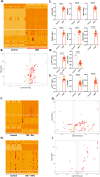
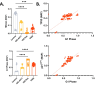
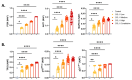
Results: Validation through simulation of classical senescence programs, including oxidative stress-induced senescence and oncogene-induced senescence, demonstrates its ability to replicate molecular signatures consistent with empirical data. Additionally, we explore the role of CCL11, a novel senescence-associated molecule, through simulations that reveal potential pathways and mechanisms underlying CCL11-mediated senescence induction.
Conclusions: In conclusion, PseudoCell provides a systematic approach to dissecting premature senescence programs and uncovering novel regulatory mechanisms.
Keywords: aging; bioinformatics; cellular senescence; in silico modeling; premature senescence.
© 2025 The Author(s). Aging Medicine published by Beijing Hospital and John Wiley & Sons Australia, Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
LinkOut - more resources
Full Text Sources
