Pathogenicity and virulence of Chlamydia trachomatis: Insights into host interactions, immune evasion, and intracellular survival
- PMID: 40353442
- PMCID: PMC12090877
- DOI: 10.1080/21505594.2025.2503423
Pathogenicity and virulence of Chlamydia trachomatis: Insights into host interactions, immune evasion, and intracellular survival
Abstract
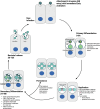
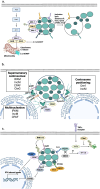
Chlamydia trachomatis is an obligate intracellular pathogen and the leading cause of bacterial sexually transmitted infections and infectious blindness worldwide. All Chlamydia species share a unique biphasic developmental cycle, alternating between infectious elementary bodies (EBs) and replicative reticulate bodies (RBs). The pathogenesis of C. trachomatis is driven by a sophisticated arsenal of adhesins, conventional type III secretion system effector proteins, and inclusion membrane proteins that subvert host cellular processes to establish infection and promote survival. In this review, we highlight the molecular mechanisms underlying C. trachomatis infection, focusing on key stages of its developmental cycle, including adhesion, invasion, replication, and egress. We delve into its interactions with host cytoskeletal structures, immune signaling pathways, and intracellular trafficking systems, as well as its strategies for immune evasion and persistence. Understanding these mechanisms offers critical insights into C. trachomatis pathogenesis and identifies promising avenues for therapeutic and vaccine development.
Keywords: Chlamydia; Inc; T3SS; adhesion; effector; host pathogen interactions.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
