Clade 2.3.4.4b highly pathogenic H5N1 influenza viruses from birds in China replicate effectively in bovine cells and pose potential public health risk
- PMID: 40353570
- PMCID: PMC12128135
- DOI: 10.1080/22221751.2025.2505649
Clade 2.3.4.4b highly pathogenic H5N1 influenza viruses from birds in China replicate effectively in bovine cells and pose potential public health risk
Abstract
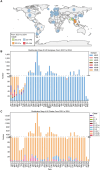
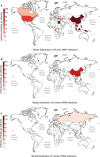
In February 2024, H5N1 highly pathogenic avian influenza viruses (HPAIVs) of clade 2.3.4.4b were first reported in dairy cows in the USA. Subsequent multiple outbreaks on dairy farms and sporadic human infections have raised substantial public health concerns. In the same year, four H5N1 HPAIVs of clade 2.3.4.4b were isolated from ducks and geese in live poultry markets (LPMs) spanning seven provinces in China. Evolutionary analysis demonstrated that these viruses had undergone two genetic reassortments with H5 influenza viruses from wild birds in different countries. Except for 565/H5N1, the other three viruses exhibited over 99% genetic homology with avian-origin H5N1 HPAIVs from South Korea and Japan. Notably, 571/H5N1 demonstrated high replication efficiency in bovine-derived cells, particularly in bovine mammary epithelial (MAC-T) cells, and caused 16.7% (1/6) mortality in mice at a dose of 105 EID50/50 μL, indicating its zoonotic potential. Given the potential cross-species transmission risk of H5N1 HPAIVs to cattle herds, we collected 228 serum samples from 12 cattle farms across five provinces and conducted serological testing to investigate seroprevalence of H5N1 HPAIVs in Chinese cattle herds. All tested samples were negative, indicating no widespread infection in the sampled cattle populations. However, infections in cattle from other regions cannot be ruled out. Nevertheless, due to the high mutability of H5N1 HPAIVs, enhanced surveillance of avian influenza viruses is critical to ensure timely responses to potential outbreaks.
Keywords: H5N1; cattle; cross-species transmission; evolution; influenza virus; zoonotic potential.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures

References
-
- Xie R, Edwards KM, Wille M, et al. The episodic resurgence of highly pathogenic avian influenza H5 virus. Nature. 2023 Oct;622(7984):810–817. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
