Mobilizable shuttle vectors with fluorescent markers functional across different species of bacteria
- PMID: 40353662
- PMCID: PMC12175534
- DOI: 10.1128/aem.00045-25
Mobilizable shuttle vectors with fluorescent markers functional across different species of bacteria
Abstract
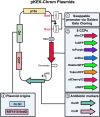
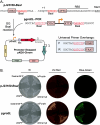
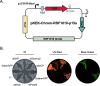
Chromophore-containing proteins (CCPs), including fluorescent and non-fluorescent (chromoproteins), have been widely used in microbiological research. However, several roadblocks often limit their use in non-model bacterial species, including efficient transformation, suitable plasmid origins of replication, and optimal promoter choice. Here, we have engineered a set of 32 shuttle plasmids designed to overcome these roadblocks in an effort to streamline this process for future research. We have selected eight different CCPs: eforCP, YukonOFP, DasherGFP, tinsel Purple, aeBlue, FuGFP, super-folder GFP, and super-folder Cherry2. To broaden the potential host range, we utilized two distinct backbones with p15a either fused to a Francisella origin (FnOri) or to the broad host origin RSF1010 and included a transfer origin (oriT) to facilitate transformation via conjugation. Moreover, we have created versions of each vector, which confer resistance to either kanamycin or chloramphenicol. Lastly, to enable promoter-swapping, we engineered the constitutive pJ23100 promoter element to be flanked by BsaI sites, thereby enabling promoter exchange by the Golden Gate assembly to evaluate CCP expression with different host promoters. To demonstrate the usability of the pKEK-Chrom plasmid series, we evaluated their expression in Escherichia coli, Shewanella oneidensis, and Vibrio alginolyticus. We further demonstrated the utility of promoter swapping in Francisella novicida and validated the functionality of the RSF1010 origin in Acinetobacter baumannii. In summary, the pKEK-Chrom plasmid series provides a palette of different CCPs that streamline their use in non-model gram-negative bacteria.
Importance: Chromophore-containing proteins (CCPs), including both fluorescent proteins and pigment-producing (non-fluorescent) chromoproteins, have become invaluable tools for microbial research. However, their successful implementation in understudied bacterial species lacking established genetic tools often requires substantial time and resources. Our goal was to develop a set of plasmid-based vectors that could streamline CCP expression in gram-negative bacteria. To do so, we developed a set of 32 plasmid vectors, the pKEK-Chrom plasmid series, specifically designed to facilitate CCP expression across different bacteria, including Escherichia coli, Vibrio alginolyticus, Shewanella oneidensis, Francisella novicida, and Acinetobacter baumannii.
Keywords: Acinetobacter baumannii; Francisella novicida; Golden Gate assembly; RSF1010; Vibrio alginolyticus; broad-host range; chromoproteins; non-model bacteria; plasmid; promoter exchange.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Liljeruhm J, Funk SK, Tietscher S, Edlund AD, Jamal S, Wistrand-Yuen P, Dyrhage K, Gynnå A, Ivermark K, Lövgren J, Törnblom V, Virtanen A, Lundin ER, Wistrand-Yuen E, Forster AC. 2018. Engineering a palette of eukaryotic chromoproteins for bacterial synthetic biology. J Biol Eng 12:8. doi: 10.1186/s13036-018-0100-0 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

