RNA sequencing analysis of viromes of Aedes albopictus and Aedes vexans collected from NEON sites
- PMID: 40354533
- PMCID: PMC12107137
- DOI: 10.1073/pnas.2403591122
RNA sequencing analysis of viromes of Aedes albopictus and Aedes vexans collected from NEON sites
Abstract
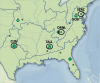
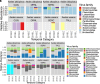
Climate change is significantly impacting the geographic range of many animal species and their associated microorganisms, hence influencing emergence of vector-borne diseases. Mosquito-borne viruses represent a potential major reservoir of human pathogens, highlighting the need for improved understanding of ecological factors associated with variation in the mosquito viral community (virome). Here, a subtractive hybridization method coupled with RNAseq of individual mosquito specimens was used to profile RNA viromes of individual co-occurring Aedes albopictus and Aedes vexans mosquitoes across a 2,000 km spatial scale. Samples were collected and archived by the National Ecological Observatory Network (NEON) from four ecologically variable sites in the Southeastern United States between 2018 and 2019. Results of multivariate analysis suggest that mosquito species are an important factor in RNA viral community composition. Significantly higher viral diversity was detected in A. albopictus compared to A.vexans. However, season, year, and site of sample collection did not show strong association with virome profiles, supporting the hypothesis that factors unique to the mosquito host species (e.g., larval habitat or vector competence) influence the structure of mosquito viromes.
Keywords: Aedes albopictus; Aedes vexans; arbovirus; mosquito; sequencing.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures



References
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

