Identification of genetic indicators linked to immunological infiltration in idiopathic pulmonary fibrosis
- PMID: 40355204
- PMCID: PMC12073941
- DOI: 10.1097/MD.0000000000042376
Identification of genetic indicators linked to immunological infiltration in idiopathic pulmonary fibrosis
Abstract
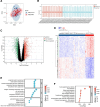
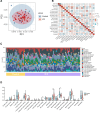
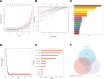
This study employed bioinformatics to investigate potential molecular markers associated with idiopathic pulmonary fibrosis (IPF) and examined their correlation with immune-infiltrating cells. Microarray data for IPF were retrieved from the Gene Expression Omnibus database. Differentially expressed genes (DEGs) and module genes were identified through Limma analysis and weighted gene co-expression network analysis. Enrichment analysis and protein-protein interaction network development were performed on the DEGs. Machine learning algorithms, including least absolute shrinkage and selection operator regression, random forest, and extreme gradient boosting, were applied to identify potential key genes. The predictive accuracy was assessed through a nomogram and a receiver operating characteristic (ROC) curve. Additionally, the correlation between core genes and immune-infiltrating cells was assessed utilizing the CIBERSORT algorithm. An IPF model was established in a human fetal lung fibroblast 1 (HFL-1) through induction with transforming growth factor β1 (TGF-β1), and validation was conducted via reverse transcription-quantitative polymerase chain reaction. A sum of 1246 genes exhibited upregulation, whereas 879 genes were downregulated. Pathway enrichment analysis and functional annotation revealed that DEGs were predominantly involved in extracellular processes. Four key genes - cd19, cxcl13, fcrl5, and slamf7 - were identified. Furthermore, ROC analysis demonstrated high predictive accuracy for these 4 genes. Compared to healthy individuals, lung tissues from IPF patients exhibited an increased presence of plasma cells, CD4 memory-activated T cells, M0 macrophages, activated dendritic cells, resting NK cells, and M2 macrophage infiltration. The upregulation of cd19, cxcl13, fcrl5, and slamf7 in TGF-β1-treated HFL-1 cells was confirmed, aligning with the findings from the microarray data analysis. cd19, cxcl13, fcrl5, and slamf7 serve as diagnostic markers for IPF, providing fresh perspectives regarding the fundamental pathogenesis and molecular mechanisms associated with this condition.
Keywords: CIBERSORT; biomarker; idiopathic pulmonary fibrosis; immune infiltration; machine learning algorithm.
Copyright © 2025 the Author(s). Published by Wolters Kluwer Health, Inc.
Conflict of interest statement
The GEO database is a publicly accessible repository that offers a comprehensive collection of ethically approved patient data. Researchers are permitted to freely download and utilize this data for scientific investigations and publication purposes. As this study was conducted using open-access datasets, no ethical concerns or conflicts of interest are associated with its findings. The authors have no funding and conflicts of interest to disclose.
Figures




Similar articles
-
Machine learning-based prediction of candidate gene biomarkers correlated with immune infiltration in patients with idiopathic pulmonary fibrosis.Front Med (Lausanne). 2023 Feb 13;10:1001813. doi: 10.3389/fmed.2023.1001813. eCollection 2023. Front Med (Lausanne). 2023. PMID: 36860337 Free PMC article.
-
Construction of an artificial neural network diagnostic model and investigation of immune cell infiltration characteristics for idiopathic pulmonary fibrosis.BMC Pulm Med. 2024 Sep 17;24(1):458. doi: 10.1186/s12890-024-03249-6. BMC Pulm Med. 2024. PMID: 39289672 Free PMC article.
-
Identifying potential biomarkers of idiopathic pulmonary fibrosis through machine learning analysis.Sci Rep. 2023 Oct 2;13(1):16559. doi: 10.1038/s41598-023-43834-z. Sci Rep. 2023. PMID: 37783761 Free PMC article.
-
Integrated bioinformatics analysis for the identification of idiopathic pulmonary fibrosis-related genes and potential therapeutic drugs.BMC Pulm Med. 2023 Oct 4;23(1):373. doi: 10.1186/s12890-023-02678-z. BMC Pulm Med. 2023. PMID: 37794454 Free PMC article.
-
Identification of oxidative stress-related diagnostic markers and immune infiltration features for idiopathic pulmonary fibrosis by bibliometrics and bioinformatics.Front Med (Lausanne). 2024 Aug 6;11:1356825. doi: 10.3389/fmed.2024.1356825. eCollection 2024. Front Med (Lausanne). 2024. PMID: 39165378 Free PMC article. Review.
References
-
- Martinez FJ, Collard HR, Pardo A, et al. . Idiopathic pulmonary fibrosis. Nat Rev Dis Primers. 2017;3:17074. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

