Spatial mapping of the brain metabolome lipidome and glycome
- PMID: 40355410
- PMCID: PMC12069719
- DOI: 10.1038/s41467-025-59487-7
Spatial mapping of the brain metabolome lipidome and glycome
Abstract
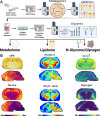
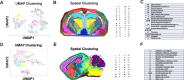
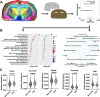
Metabolites, lipids, and glycans are fundamental but interconnected classes of biomolecules that form the basis of the metabolic network. These molecules are dynamically channeled through multiple pathways that govern cellular physiology and pathology. Here, we present a framework for the simultaneous spatial analysis of the metabolome, lipidome, and glycome from a single tissue section using mass spectrometry imaging. This workflow integrates a computational platform, the Spatial Augmented Multiomics Interface (Sami), which enables multiomics integration, high-dimensional clustering, spatial anatomical mapping of matched molecular features, and metabolic pathway enrichment. To demonstrate the utility of this approach, we applied Sami to evaluate metabolic diversity across distinct brain regions and to compare wild-type and Ps19 Alzheimer's disease (AD) mouse models. Our findings reveal region-specific metabolic demands in the normal brain and highlight metabolic dysregulation in the Ps19 model, providing insights into the biochemical alterations associated with neurodegeneration.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: R.C.S. has research support and received consultancy fees from Maze Therapeutics. R.C.S. is a member of the Medical Advisory Board for Little Warrior Foundation and M.S.G. is a member of the science advisory board for Chelsea’s Hope, Glut1-deficiency syndrome, and the adult polyglucosan body disease foundation. M.S.G. has research support and research compounds from Maze Therapeutics, Valerion Therapeutics, Ionis Pharmaceuticals. M.S.G. also received consultant fees from Maze Therapeutics. The remaining authors declare no competing interests.
Figures


Update of
-
Spatial Metabolome Lipidome and Glycome from a Single brain Section.bioRxiv [Preprint]. 2023 Jul 25:2023.07.22.550155. doi: 10.1101/2023.07.22.550155. bioRxiv. 2023. Update in: Nat Commun. 2025 May 12;16(1):4373. doi: 10.1038/s41467-025-59487-7. PMID: 37546843 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

