Pharmacological inhibition of PLK1/PRC1 triggers mitotic catastrophe and sensitizes lung cancers to chemotherapy
- PMID: 40355412
- PMCID: PMC12069692
- DOI: 10.1038/s41419-025-07708-8
Pharmacological inhibition of PLK1/PRC1 triggers mitotic catastrophe and sensitizes lung cancers to chemotherapy
Abstract
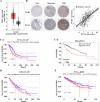
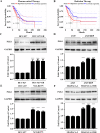
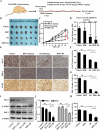
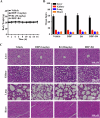
Polo-like kinase 1 (PLK1) signaling drives tumor malignancy and chemotherapy resistance, which is an unmet clinical need. Recruiting PLK1 to the central spindle during anaphase is necessary for its function in promoting cancer cell proliferation, which is achieved by binding to microtubule-associated protein regulating of cytokinesis (PRC1) located in the spindle. However, the role of PLK1/PRC1 signaling in chemotherapy resistance is unknown. In this study, we identified a small molecule B4 which inhibited PLK1/PRC1 signaling through disrupting the formation of PLK1/PRC1 protein complexes. In the presence of blocking PLK1/PRC1 signaling, enhanced sensitivity of drug-resistant tumors to traditional chemotherapy was found. Suppression of PLK1 activity by B4 inhibited disease progression in allograft models, and combination with cisplatin elicited dramatic regression of drug-resistant tumors. Our findings provide a promising strategy to target the PLK1 signaling cascade and demonstrate a potential modality to enhance sensitivity to chemotherapy in non-small cell lung cancer (NSCLC).
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

