Twin pair analysis uncovers links between DNA methylation, mitochondrial DNA quantity and obesity
- PMID: 40355419
- PMCID: PMC12069627
- DOI: 10.1038/s41467-025-59576-7
Twin pair analysis uncovers links between DNA methylation, mitochondrial DNA quantity and obesity
Abstract
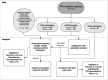
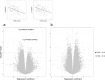
Alterations in mitochondrial metabolism in obesity may indicate disrupted communication between mitochondria and nucleus, and DNA methylation may influence this interplay. Here, we leverage data from the Finnish Twin Cohort study subcohort (n = 173; 86 full twin pairs, 1 singleton), including comprehensive measurements of obesity-related outcomes, mitochondrial DNA quantity and nuclear DNA methylation levels in adipose and muscle tissue, to identify one CpG at SH3BP4 significantly associated with mitochondrial DNA quantity in adipose tissue (FDR < 0.05). We also show that SH3BP4 methylation correlates with its gene expression. Additionally, we find that 14 out of the 35 obesity-related traits display significant associations with both SH3BP4 methylation and mitochondrial DNA quantity in adipose tissue. We use data from TwinsUK and the Scandinavian T2D-discordant monozygotic twin cohort, to validate the observed associations. Further analysis using ICE FALCON suggests that mitochondrial DNA quantity, insulin sensitivity and certain body fat measures are causal to SH3BP4 methylation. Examining mitochondrial DNA quantity and obesity-related traits suggests causation from mitochondrial DNA quantity to obesity, but unmeasured within-individual confounding cannot be ruled out. Our findings underscore the impact of mitochondrial DNA quantity on DNA methylation and expression of the SH3BP4 gene within adipose tissue, with potential implications for obesity.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

