Multi-omic analysis reveals a CAF-stemness-governed classification in HCC liver transplant recipients beyond the Milan criteria
- PMID: 40355422
- PMCID: PMC12069600
- DOI: 10.1038/s41467-025-59745-8
Multi-omic analysis reveals a CAF-stemness-governed classification in HCC liver transplant recipients beyond the Milan criteria
Abstract
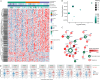
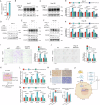
In patients with hepatocellular carcinoma (HCC) meeting the Milan criteria, liver transplantation (LT) is an effective therapy. This study aims to define the survival-related molecular biological features helping precisely identifying the patients with HCC beyond the Milan criteria who have acceptable outcomes. In the derivation cohort, integrated analyses of tumor tissues are conducted using RNA sequencing (RNA-seq), proteomic landscape, and transposase-accessible chromatin sequencing (ATAC-seq). Based on transcriptomics, three subgroups that significantly differ in overall survival were identified in the derivation cohort, and these findings are validated in an independent cohort. In-depth bioinformatics analysis using RNA-seq and proteomics reveals that the promotion of cancer stemness by cancer-associated fibroblasts (CAFs) can be responsible for the negative biological characteristics observed in high-risk HCC patients. The ATAC-seq identifies key factors regulating transcription, which may bridge CAF infiltration and stemness. Finally, we demonstrate that the CAF-derived CXCL12 sustains the stemness of HCC cells by promoting XRCC5 through CXCR4.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Ethics approval statement: The study protocol was approved by the Review Boards of the NHC Key Laboratory of Combined Multi-organ Transplantation. The Ethical review approval number: 2018-768.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

