Comprehensive molecular profiling of FH-deficient renal cell carcinoma identifies molecular subtypes and potential therapeutic targets
- PMID: 40355427
- PMCID: PMC12069531
- DOI: 10.1038/s41467-025-59513-8
Comprehensive molecular profiling of FH-deficient renal cell carcinoma identifies molecular subtypes and potential therapeutic targets
Abstract
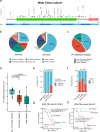
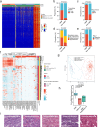
Fumarate hydratase-deficient renal cell carcinoma (FH-deficient RCC) is a rare yet highly lethal kidney cancer. To deepen our understanding of FH-deficient RCC, we conduct a comprehensive integrated genomic study. We analyze the association of FH alteration patterns with tumor heterogeneity and develop a CpG site-specific methylation signature for precise identification of FH-deficient RCC. Transcriptomic analysis unveils three distinctive molecular subtypes characterized by enrichment of immune/Angiogenic/Stromal (C1), WNT/Notch/MAPK (C2), and proliferation/stemness (C3) pathways, respectively. Tumors in C1 derive the most substantial survival benefit from a combination of immune checkpoint blockade (ICB) and anti-angiogenic therapy. Tumors in C2 display moderate response to this therapeutic approach. In contrast, tumors in C3 exhibit an unfavorable response to anti-angiogenic monotherapy and its combination with ICB. These findings contribute to a profound understanding of the aggressive nature of FH-deficient RCC, offering insights into potential precision medicine approaches for disease management.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




References
-
- Gardie, B. et al. Novel FH mutations in families with hereditary leiomyomatosis and renal cell cancer (HLRCC) and patients with isolated type 2 papillary renal cell carcinoma. J. Med. Genet.48, 226–234 (2011). - PubMed
-
- Ashrafian, H. et al. Expression profiling in progressive stages of fumarate-hydratase deficiency: the contribution of metabolic changes to tumorigenesis. Cancer Res.70, 9153–9165 (2010). - PubMed
MeSH terms
Substances
Grants and funding
- NSFC 82472673/National Natural Science Foundation of China (National Science Foundation of China)
- NSFC 82403766/National Natural Science Foundation of China (National Science Foundation of China)
- NSFC 82103097/National Natural Science Foundation of China (National Science Foundation of China)
- NSFC 81872107/National Natural Science Foundation of China (National Science Foundation of China)
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

