ALDH4A1 functions as an active component of the MPC complex maintaining mitochondrial pyruvate import for TCA cycle entry and tumour suppression
- PMID: 40355545
- PMCID: PMC12188464
- DOI: 10.1038/s41556-025-01651-8
ALDH4A1 functions as an active component of the MPC complex maintaining mitochondrial pyruvate import for TCA cycle entry and tumour suppression
Abstract
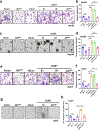
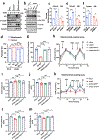
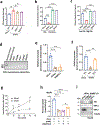
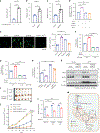
MPC1 and MPC2 are two well-known components of the mitochondrial pyruvate carrier (MPC) complex maintaining MPC activity to transport pyruvate into mitochondria for tricarboxylic acid (TCA) cycle entry in mammalian cells. It is currently unknown whether there is an additional MPC component crucially maintaining MPC complex activity for pyruvate mitochondrial import. Here we show that ALDH4A1, a proline-metabolizing enzyme localized in mitochondria, serves as a previously unrecognized MPC component maintaining pyruvate mitochondrial import and the TCA cycle independently of its enzymatic activity. Loss of ALDH4A1 in mammalian cells impairs pyruvate entry to mitochondria, resulting in defective TCA cycle entry. ALDH4A1 forms an active trimeric complex with MPC1-MPC2 to maintain the integrity and oligomerization of MPC1-MPC2 and facilitates pyruvate transport in an in vitro system. ALDH4A1 displays tumour suppression by maintaining MPC complex activity. Our study identifies ALDH4A1 as an essential component of MPC for pyruvate mitochondrial import, TCA cycle entry and tumour suppression.
© 2025. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests: H.K.L. is a consultant for Stablix, Inc. and Chang Gung University of Science and Technology. All other authors declare no competing interests.
Figures














References
MeSH terms
Substances
Grants and funding
- R01CA248037/U.S. Department of Health & Human Services | NIH | NCI | Division of Cancer Epidemiology and Genetics, National Cancer Institute (National Cancer Institute Division of Cancer Epidemiology and Genetics)
- R01CA270617/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- P30 CA012197/CA/NCI NIH HHS/United States
- R01 CA256158/CA/NCI NIH HHS/United States
- R01CA256158/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

